3dSS: 3D structural superposition
- PMID: 16844975
- PMCID: PMC1538824
- DOI: 10.1093/nar/gkl036
3dSS: 3D structural superposition
Abstract
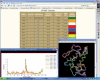
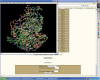
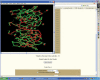
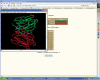
3dSS is a web-based interactive computing server, primarily designed to aid researchers, to superpose two or several 3D protein structures. In addition, the server can be effectively used to find the invariant and common water molecules present in the superposed homologous protein structures. The molecular visualization tool RASMOL is interfaced with the server to visualize the superposed 3D structures with the water molecules (invariant or common) in the client machine. Furthermore, an option is provided to save the superposed 3D atomic coordinates in the client machine. To perform the above, users need to enter Protein Data Bank (PDB)-id(s) or upload the atomic coordinates in PDB format. This server uses a locally maintained PDB anonymous FTP server that is being updated weekly. This program can be accessed through our Bioinformatics web server at the URL http://cluster.physics.iisc.ernet.in/3dss/ or http://10.188.1.15/3dss/.
Figures
References
-
- Bernstein F.C., Koetzle T.F., Williams G.J.B., Meyer E.F., Jr, Brice M.D., Rogers J.R., Kennard O., Shimanouchi T., Tasumi M.J. The Protein Data Bank: a computer based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
-
- Kabsch W.A. Discussion of solution for best rotation of two vectors. Acta Crystallogr. 1978;A34:827–828.
-
- MacLachlan A.D. Rapid comparison of protein structures. Acta Crystallogr. 1982;A38:871–873.
-
- Kearsley S.K. An algorithm for the simultaneous superposition of a structural series. J. Comput. Chem. 1990;11:1187–1192.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

