AlgPred: prediction of allergenic proteins and mapping of IgE epitopes
- PMID: 16844994
- PMCID: PMC1538830
- DOI: 10.1093/nar/gkl343
AlgPred: prediction of allergenic proteins and mapping of IgE epitopes
Abstract
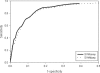
In this study a systematic attempt has been made to integrate various approaches in order to predict allergenic proteins with high accuracy. The dataset used for testing and training consists of 578 allergens and 700 non-allergens obtained from A. K. Bjorklund, D. Soeria-Atmadja, A. Zorzet, U. Hammerling and M. G. Gustafsson (2005) Bioinformatics, 21, 39-50. First, we developed methods based on support vector machine using amino acid and dipeptide composition and achieved an accuracy of 85.02 and 84.00%, respectively. Second, a motif-based method has been developed using MEME/MAST software that achieved sensitivity of 93.94 with 33.34% specificity. Third, a database of known IgE epitopes was searched and this predicted allergenic proteins with 17.47% sensitivity at specificity of 98.14%. Fourth, we predicted allergenic proteins by performing BLAST search against allergen representative peptides. Finally hybrid approaches have been developed, which combine two or more than two approaches. The performance of all these algorithms has been evaluated on an independent dataset of 323 allergens and on 101 725 non-allergens obtained from Swiss-Prot. A web server AlgPred has been developed for the predicting allergenic proteins and for mapping IgE epitopes on allergenic proteins (http://www.imtech.res.in/raghava/algpred/). AlgPred is available at www.imtech.res.in/raghava/algpred/.
Figures
Similar articles
-
AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes.Brief Bioinform. 2021 Jul 20;22(4):bbaa294. doi: 10.1093/bib/bbaa294. Brief Bioinform. 2021. PMID: 33201237
-
A machine learning based method for the prediction of secretory proteins using amino acid composition, their order and similarity-search.In Silico Biol. 2008;8(2):129-40. In Silico Biol. 2008. PMID: 18928201
-
CEP: a conformational epitope prediction server.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W168-71. doi: 10.1093/nar/gki460. Nucleic Acids Res. 2005. PMID: 15980448 Free PMC article.
-
Evaluation of available IgE-binding epitope data and its utility in bioinformatics.Mol Nutr Food Res. 2006 Jul;50(7):638-44. doi: 10.1002/mnfr.200500276. Mol Nutr Food Res. 2006. PMID: 16764019 Review.
-
Practical and predictive bioinformatics methods for the identification of potentially cross-reactive protein matches.Mol Nutr Food Res. 2006 Jul;50(7):655-60. doi: 10.1002/mnfr.200500277. Mol Nutr Food Res. 2006. PMID: 16810734 Review.
Cited by
-
Structural Basis for Designing Multiepitope Vaccines Against COVID-19 Infection: In Silico Vaccine Design and Validation.JMIR Bioinform Biotechnol. 2020 Jun 19;1(1):e19371. doi: 10.2196/19371. eCollection 2020 Jan-Dec. JMIR Bioinform Biotechnol. 2020. PMID: 32776022 Free PMC article.
-
Brachypodium distachyon as a model for defining the allergen potential of non-prolamin proteins.Funct Integr Genomics. 2012 Aug;12(3):439-46. doi: 10.1007/s10142-012-0294-z. Epub 2012 Aug 30. Funct Integr Genomics. 2012. PMID: 22933233 Free PMC article.
-
Benzo[e]pyrimido[5,4-b][1,4]diazepin-6(11H)-one derivatives as Aurora A kinase inhibitors: LQTA-QSAR analysis and detailed systematic validation of the developed model.Mol Divers. 2015 Nov;19(4):965-74. doi: 10.1007/s11030-015-9618-y. Epub 2015 Jul 17. Mol Divers. 2015. PMID: 26183841
-
Mining of Marburg Virus Proteome for Designing an Epitope-Based Vaccine.Front Immunol. 2022 Jul 15;13:907481. doi: 10.3389/fimmu.2022.907481. eCollection 2022. Front Immunol. 2022. PMID: 35911751 Free PMC article.
-
Towards the First Multiepitope Vaccine Candidate against Neospora caninum in Mouse Model: Immunoinformatic Standpoint.Biomed Res Int. 2022 Jun 9;2022:2644667. doi: 10.1155/2022/2644667. eCollection 2022. Biomed Res Int. 2022. PMID: 35722460 Free PMC article.
References
-
- Sadok Y. Diagnosis of type I hypersensitivity by laboratory test. Tunis Med. 2005;83:441–444. - PubMed
-
- Scheurer S., Son D.Y., Boehm M., Karamloo F., Franke S., Hoffmann A., Haustein D., Vieths S. Cross-reactivity and epitope analysis of Pru a 1, the major cherry allergen. Mol. Immunol. 1999;36:155–167. - PubMed
-
- Santos A.B., Chapman M.D., Aalberse R.C., Vailes L.D., Ferriani V.P., Oliver C., Rizzo M.C., Naspitz C.K., Arruda L.K. Cockroach allergens and asthma in Brazil: identification of tropomyosin as a major allergen with potential cross-reactivity with mite and shrimp allergens. J. Allergy Clin. Immunol. 1999;104:329–337. - PubMed
-
- Broadfield E., McKeever T.M., Scrivener S., Venn A., Lewis S.A., Britton J. Increase in the prevalence of allergen skin sensitization in successive birth cohorts. J. Allergy Clin. Immunol. 2002;109:969–974. - PubMed
-
- Goodman R.E., Hefle S.L., Taylor S.L., Ree R.V. Assessing genetically modified crops to minimize the risk of increased food allergy: a review. Int. Arch. Allergy Immunol. 2005;137:153–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials