MAGIIC-PRO: detecting functional signatures by efficient discovery of long patterns in protein sequences
- PMID: 16845025
- PMCID: PMC1538832
- DOI: 10.1093/nar/gkl309
MAGIIC-PRO: detecting functional signatures by efficient discovery of long patterns in protein sequences
Erratum in
- Nucleic Acids Res. 2008 Mar;36(4):1400-6
Corrected and republished in
-
MAGIIC-PRO: detecting functional signatures by efficient discovery of long patterns in protein sequences.Nucleic Acids Res. 2008 Mar;36(4):1400-6. doi: 10.1093/nar/gkm717. Nucleic Acids Res. 2008. PMID: 18314547 Free PMC article.
Abstract
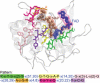
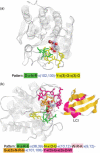
This paper presents a web service named MAGIIC-PRO, which aims to discover functional signatures of a query protein by sequential pattern mining. Automatic discovery of patterns from unaligned biological sequences is an important problem in molecular biology. MAGIIC-PRO is different from several previously established methods performing similar tasks in two major ways. The first remarkable feature of MAGIIC-PRO is its efficiency in delivering long patterns. With incorporating a new type of gap constraints and some of the state-of-the-art data mining techniques, MAGIIC-PRO usually identifies satisfied patterns within an acceptable response time. The efficiency of MAGIIC-PRO enables the users to quickly discover functional signatures of which the residues are not from only one region of the protein sequences or are only conserved in few members of a protein family. The second remarkable feature of MAGIIC-PRO is its effort in refining the mining results. Considering large flexible gaps improves the completeness of the derived functional signatures. The users can be directly guided to the patterns with as many blocks as that are conserved simultaneously. In this paper, we show by experiments that MAGIIC-PRO is efficient and effective in identifying ligand-binding sites and hot regions in protein-protein interactions directly from sequences. The web service is available at http://biominer.bime.ntu.edu.tw/magiicpro and a mirror site at http://biominer.cse.yzu.edu.tw/magiicpro.
Figures

Similar articles
-
Identification of hot regions in protein-protein interactions by sequential pattern mining.BMC Bioinformatics. 2007 May 24;8 Suppl 5(Suppl 5):S8. doi: 10.1186/1471-2105-8-S5-S8. BMC Bioinformatics. 2007. PMID: 17570867 Free PMC article.
-
iPDA: integrated protein disorder analyzer.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W465-72. doi: 10.1093/nar/gkm353. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553839 Free PMC article.
-
WildSpan: mining structured motifs from protein sequences.Algorithms Mol Biol. 2011 Mar 31;6(1):6. doi: 10.1186/1748-7188-6-6. Algorithms Mol Biol. 2011. PMID: 21453542 Free PMC article.
-
MAGIIC-PRO: detecting functional signatures by efficient discovery of long patterns in protein sequences.Nucleic Acids Res. 2008 Mar;36(4):1400-6. doi: 10.1093/nar/gkm717. Nucleic Acids Res. 2008. PMID: 18314547 Free PMC article.
-
Discovering patterns and subfamilies in biosequences.Proc Int Conf Intell Syst Mol Biol. 1996;4:34-43. Proc Int Conf Intell Syst Mol Biol. 1996. PMID: 8877502 Review.
Cited by
-
Identification of hot regions in protein-protein interactions by sequential pattern mining.BMC Bioinformatics. 2007 May 24;8 Suppl 5(Suppl 5):S8. doi: 10.1186/1471-2105-8-S5-S8. BMC Bioinformatics. 2007. PMID: 17570867 Free PMC article.
-
iPDA: integrated protein disorder analyzer.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W465-72. doi: 10.1093/nar/gkm353. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553839 Free PMC article.
-
WildSpan: mining structured motifs from protein sequences.Algorithms Mol Biol. 2011 Mar 31;6(1):6. doi: 10.1186/1748-7188-6-6. Algorithms Mol Biol. 2011. PMID: 21453542 Free PMC article.
-
E1DS: catalytic site prediction based on 1D signatures of concurrent conservation.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W291-6. doi: 10.1093/nar/gkn324. Epub 2008 Jun 4. Nucleic Acids Res. 2008. PMID: 18524800 Free PMC article.
-
Predicting DNA-binding locations and orientation on proteins using knowledge-based learning of geometric properties.Proteome Sci. 2011 Oct 14;9 Suppl 1(Suppl 1):S11. doi: 10.1186/1477-5956-9-S1-S11. Proteome Sci. 2011. PMID: 22166082 Free PMC article.
References
-
- Saqi M.A.S., Sternberg M.J.E. Identification of sequence motifs from a set of proteins with related function. Protein Eng. 1994;7:165–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

