HHsenser: exhaustive transitive profile search using HMM-HMM comparison
- PMID: 16845029
- PMCID: PMC1538784
- DOI: 10.1093/nar/gkl195
HHsenser: exhaustive transitive profile search using HMM-HMM comparison
Abstract
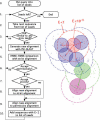
HHsenser is the first server to offer exhaustive intermediate profile searches, which it combines with pairwise comparison of hidden Markov models. Starting from a single protein sequence or a multiple alignment, it can iteratively explore whole superfamilies, producing few or no false positives. The output is a multiple alignment of all detected homologs. HHsenser's sensitivity should make it a useful tool for evolutionary studies. It may also aid applications that rely on diverse multiple sequence alignments as input, such as homology-based structure and function prediction, or the determination of functional residues by conservation scoring and functional subtyping.HHsenser can be accessed at http://hhsenser.tuebingen.mpg.de/. It has also been integrated into our structure and function prediction server HHpred (http://hhpred.tuebingen.mpg.de/) to improve predictions for near-singleton sequences.
Figures

References
-
- Przybylski D., Rost B. Alignments grow, secondary structure prediction improves. Proteins. 2002;46:197–205. - PubMed
-
- Garg A., Kaur H., Raghava G.P. Real value prediction of solvent accessibility in proteins using multiple sequence alignment and secondary structure. Proteins. 2005;61:318–324. - PubMed
-
- Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005;3:35–60. - PubMed
-
- Punta M., Rost B. PROFcon: novel prediction of long-range contacts. Bioinformatics. 2005;21:2960–2968. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

