CrossLink: visualization and exploration of sequence relationships between (micro) RNAs
- PMID: 16845036
- PMCID: PMC1538911
- DOI: 10.1093/nar/gkl223
CrossLink: visualization and exploration of sequence relationships between (micro) RNAs
Abstract
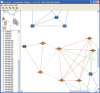
CrossLink is a versatile tool for the exploration of relationships between RNA sequences. After a parametrization phase, CrossLink delegates the determination of sequence relationships to established tools (BLAST, Vmatch and RNAhybrid) and then constructs a network. Each node in this network represents a sequence and each link represents a match or a set of matches. Match attributes are reflected by graphical attributes of the links and corresponding alignments are displayed on a mouse-click. The distributions of match attributes such as E-value, match length and proportion of identical nucleotides are displayed as histograms. Sequence sets can be highlighted and visibility of designated matches can be suppressed by real-time adjustable thresholds for attribute combinations. Powerful network layout operations (such as spring-embedding algorithms) and navigation capabilities complete the exploration features of this tool. CrossLink can be especially useful in a microRNA context since Vmatch and RNAhybrid are suitable tools for determining the antisense and hybridization relationships, which are decisive for the interaction between microRNAs and their targets. CrossLink is available both online and as a standalone version at http://www-ab.informatik.uni-tuebingen.de/software.
Figures



References
-
- Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

