ASPIC: a web resource for alternative splicing prediction and transcript isoforms characterization
- PMID: 16845044
- PMCID: PMC1538898
- DOI: 10.1093/nar/gkl324
ASPIC: a web resource for alternative splicing prediction and transcript isoforms characterization
Abstract
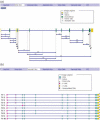
Alternative splicing (AS) is now emerging as a major mechanism contributing to the expansion of the transcriptome and proteome complexity of multicellular organisms. The fact that a single gene locus may give rise to multiple mRNAs and protein isoforms, showing both major and subtle structural variations, is an exceptionally versatile tool in the optimization of the coding capacity of the eukaryotic genome. The huge and continuously increasing number of genome and transcript sequences provides an essential information source for the computational detection of genes AS pattern. However, much of this information is not optimally or comprehensively used in gene annotation by current genome annotation pipelines. We present here a web resource implementing the ASPIC algorithm which we developed previously for the investigation of AS of user submitted genes, based on comparative analysis of available transcript and genome data from a variety of species. The ASPIC web resource provides graphical and tabular views of the splicing patterns of all full-length mRNA isoforms compatible with the detected splice sites of genes under investigation as well as relevant structural and functional annotation. The ASPIC web resource-available at http://www.caspur.it/ASPIC/--is dynamically interconnected with the Ensembl and Unigene databases and also implements an upload facility.
Figures


References
-
- Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell Biol. 2005;6:386–398. - PubMed
-
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature. 2002;420:563–573. - PubMed
-
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
-
- Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

