PROTOGENE: turning amino acid alignments into bona fide CDS nucleotide alignments
- PMID: 16845080
- PMCID: PMC1538918
- DOI: 10.1093/nar/gkl170
PROTOGENE: turning amino acid alignments into bona fide CDS nucleotide alignments
Abstract
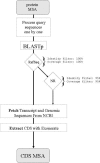
We describe Protogene, a server that can turn a protein multiple sequence alignment into the equivalent alignment of the original gene coding DNA. Protogene relies on a pipeline where every initial protein sequence is BLASTed against RefSeq or NR. The annotation associated with potential matches is used to identify the gene sequence. This gene sequence is then aligned with the query protein using Exonerate in order to extract a coding nucleotide sequence matching the original protein. Protogene can handle protein fragments and will return every CDS coding for a given protein, even if they occur in different genomes. Protogene is available from http://www.tcoffee.org/.
Figures


References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

