Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee
- PMID: 16845081
- PMCID: PMC1538866
- DOI: 10.1093/nar/gkl092
Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee
Abstract
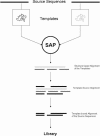
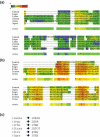
Expresso is a multiple sequence alignment server that aligns sequences using structural information. The user only needs to provide sequences. The server runs BLAST to identify close homologues of the sequences within the PDB database. These PDB structures are used as templates to guide the alignment of the original sequences using structure-based sequence alignment methods like SAP or Fugue. The final result is a multiple sequence alignment of the original sequences based on the structural information of the templates. An advanced mode makes it possible to either upload private structures or specify which PDB templates should be used to model each sequence. Providing the suitable structural information is available, Expresso delivers sequence alignments with accuracy comparable with structure-based alignments. The server is available on http://www.tcoffee.org/.
Figures
References
-
- Sander C., Schneider R. Database of homology-derived structures and the structurally meaning of sequence alignment. Proteins. 1991;9:56–68. - PubMed
-
- Holm L., Sander C. Mapping the protein universe. Science. 1996;283:595–602. - PubMed
-
- Lesk A.M., Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol. 1980;136:225–270. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

