New Onto-Tools: Promoter-Express, nsSNPCounter and Onto-Translate
- PMID: 16845086
- PMCID: PMC1538776
- DOI: 10.1093/nar/gkl213
New Onto-Tools: Promoter-Express, nsSNPCounter and Onto-Translate
Abstract
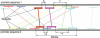
The Onto-Tools suite is composed of an annotation database and eight complementary, web-accessible data mining tools: Onto-Express, Onto-Compare, Onto-Design, Onto-Translate, Onto-Miner, Pathway-Express, Promoter-Express and nsSNPCounter. Promoter-Express is a new tool added to the Onto-Tools ensemble that facilitates the identification of transcription factor binding sites active in specific conditions. nsSNPCounter is another new tool that allows computation and analysis of synonymous and non-synonymous codon substitutions for studying evolutionary rates of protein coding genes. Onto-Translate has also been enhanced to expand its scope and accuracy by fully utilizing the capabilities of the Onto-Tools database. Currently, Onto-Translate allows arbitrary mappings between 28 types of IDs for 53 organisms. Onto-Tools are freely available at http://vortex.cs.wayne.edu/Projects.html.
Figures


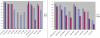
Similar articles
-
Onto-Tools: new additions and improvements in 2006.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W206-11. doi: 10.1093/nar/gkm327. Epub 2007 Jun 21. Nucleic Acids Res. 2007. PMID: 17584796 Free PMC article.
-
Recent additions and improvements to the Onto-Tools.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W762-5. doi: 10.1093/nar/gki472. Nucleic Acids Res. 2005. PMID: 15980579 Free PMC article.
-
Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W449-56. doi: 10.1093/nar/gkh409. Nucleic Acids Res. 2004. PMID: 15215428 Free PMC article.
-
Biomolecular pathway databases.Methods Mol Biol. 2010;609:129-44. doi: 10.1007/978-1-60327-241-4_8. Methods Mol Biol. 2010. PMID: 20221917 Review.
-
rSNP_Guide: an integrated database-tools system for studying SNPs and site-directed mutations in transcription factor binding sites.Hum Mutat. 2002 Oct;20(4):239-48. doi: 10.1002/humu.10116. Hum Mutat. 2002. PMID: 12325018 Review.
Cited by
-
Onto-Tools: new additions and improvements in 2006.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W206-11. doi: 10.1093/nar/gkm327. Epub 2007 Jun 21. Nucleic Acids Res. 2007. PMID: 17584796 Free PMC article.
-
Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14.Nat Med. 2011 Jan;17(1):96-104. doi: 10.1038/nm.2270. Epub 2010 Dec 12. Nat Med. 2011. PMID: 21151137 Free PMC article.
-
Inflammatory gene regulatory networks in amnion cells following cytokine stimulation: translational systems approach to modeling human parturition.PLoS One. 2011;6(6):e20560. doi: 10.1371/journal.pone.0020560. Epub 2011 Jun 2. PLoS One. 2011. PMID: 21655103 Free PMC article.
-
Gene expression profiling reveals signatures characterizing histologic subtypes of hepatoblastoma and global deregulation in cell growth and survival pathways.Hum Pathol. 2009 Jun;40(6):843-53. doi: 10.1016/j.humpath.2008.10.022. Epub 2009 Feb 5. Hum Pathol. 2009. PMID: 19200578 Free PMC article.
-
DDEC: Dragon database of genes implicated in esophageal cancer.BMC Cancer. 2009 Jul 6;9:219. doi: 10.1186/1471-2407-9-219. BMC Cancer. 2009. PMID: 19580656 Free PMC article.
References
-
- Drăghici S., Khatri P., Martins R.P., Ostermeier G.C., Krawetz S.A. Global functional profiling of gene expression. Genomics. 2003;81:98–104. - PubMed
-
- Drăghici S., Khatri P., Shah A., Tainsky M. Assessing the functional bias of commercial microarrays using the Onto-Compare database. BioTechniques. 2003:55–61. - PubMed
-
- Khatri P., Drăghici S., Ostermeier G.C., Krawetz S.A. Profiling gene expression using Onto-Express. Genomics. 2002;79:266–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

