FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization
- PMID: 16845089
- PMCID: PMC1538865
- DOI: 10.1093/nar/gkl236
FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization
Abstract
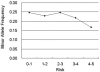
Single nucleotide polymorphism (SNP) prioritization based on the phenotypic risk is essential for association studies. Assessment of the risk requires access to a variety of heterogeneous biological databases and analytical tools. FASTSNP (function analysis and selection tool for single nucleotide polymorphisms) is a web server that allows users to efficiently identify and prioritize high-risk SNPs according to their phenotypic risks and putative functional effects. A unique feature of FASTSNP is that the functional effect information used for SNP prioritization is always up-to-date, because FASTSNP extracts the information from 11 external web servers at query time using a team of web wrapper agents. Moreover, FASTSNP is extendable by simply deploying more Web wrapper agents. To validate the results of our prioritization, we analyzed 1569 SNPs from the SNP500Cancer database. The results show that SNPs with a high predicted risk exhibit low allele frequencies for the minor alleles, consistent with a well-known finding that a strong selective pressure exists for functional polymorphisms. We have been using FASTSNP for 2 years and FASTSNP enables us to discover a novel promoter polymorphism. FASTSNP is available at http://fastsnp.ibms.sinica.edu.tw.
Figures
References
-
- Tabor H.K., Risch N.J., Myers R.M. Opinion: candidate-gene approaches for studying complex genetic traits: practical considerations. Nature Rev. Genet. 2002;3:391–397. - PubMed
-
- Hsu C.-N., Chang C.-H., Siek H., Lu J.-J., Chiou J.-J. Reconfigurable web wrapper agents for web information integration. IEEE Intell. Syst. 2003;18:34–40.
-
- Hsu C.-N., Chang C.-H., Hsieh C.-H., Lu J.-J., Chang C.-C. Reconfigurable Web wrapper agents for biological information integration. J. Am. Soc. Info. Sci. Tech. 2005;56:505–517.
-
- TheInternationalHapMapConsortium. The International HapMap Project. Nature. 2003;426:789–796. - PubMed
-
- Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C.R., Lim E.P., Kalyanaraman N., et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 1999;22:231–238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

