CorGen--measuring and generating long-range correlations for DNA sequence analysis
- PMID: 16845099
- PMCID: PMC1538783
- DOI: 10.1093/nar/gkl234
CorGen--measuring and generating long-range correlations for DNA sequence analysis
Abstract
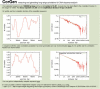
CorGen is a web server that measures long-range correlations in the base composition of DNA and generates random sequences with the same correlation parameters. Long-range correlations are characterized by a power-law decay of the auto correlation function of the GC-content. The widespread presence of such correlations in eukaryotic genomes calls for their incorporation into accurate null models of eukaryotic DNA in computational biology. For example, the score statistics of sequence alignment and the performance of motif finding algorithms are significantly affected by the presence of genomic long-range correlations. We use an expansion-randomization dynamics to efficiently generate the correlated random sequences. The server is available at http://corgen.molgen.mpg.de.
Figures
References
-
- Peng C.-K., Buldyrev S.V., Goldberger A.L., Havlin S., Sciortino F., Simons M., Stanley H.E. Long-range correlations in nucleotide sequences. Nature. 1992;356:168. - PubMed
-
- Li W., Kaneko K. Long-range correlation and partial 1/fα spectrum in a noncoding DNA sequence. Europhys. Lett. 1992;17:655.
-
- Voss R.F. Evolution of long-range fractal correlations and 1/f noise in DNA base sequences. Phys. Rev. Lett. 1992;68:3805. - PubMed
-
- Arneodo A., Bacry E., Graves P.V., Muzy J.F. Characterizing long-range correlations in DNA sequences from wavelet analysis. Phys. Rev. Lett. 1995;74:3293. - PubMed
-
- Bernaola-Galvan P., Carpena P., Roman-Roldan R., Oliver J.L. Study of statistical correlations in DNA sequences. Gene. 2002;300:105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

