FAF-Drugs: free ADME/tox filtering of compound collections
- PMID: 16845110
- PMCID: PMC1538885
- DOI: 10.1093/nar/gkl065
FAF-Drugs: free ADME/tox filtering of compound collections
Abstract
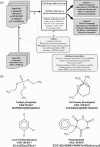
In silico screening based on the structures of the ligands or of the receptors has become an essential tool to facilitate the drug discovery process but compound collections are needed to carry out such in silico experiments. It has been recognized that absorption, distribution, metabolism, excretion and toxicity (ADME/tox) are key properties that need to be considered early on, even during the database preparation stage. FAF-Drugs is an online service based on Frowns (a chemoinformatics toolkit) that allows users to process their own compound collections via simple ADME/Tox filtering rules such as molecular weight, polar surface area, logP or number of rotatable bonds. SMILES (Simplified Molecular Input Line Entry System), CANSMILES (canonical smiles) or SDF (structure data file) files are required as input and molecules that pass or do not pass the filters are sent back in CANSMILES format. This service should thus help scientists engaging in drug discovery campaigns. Other utilities and several compound collections suitable for in silico screening are available at our site. FAF-Drugs can be accessed at http://bioserv.rpbs.jussieu.fr/FAFDrugs.html.
Figures

Similar articles
-
FAF-Drugs2: free ADME/tox filtering tool to assist drug discovery and chemical biology projects.BMC Bioinformatics. 2008 Sep 24;9:396. doi: 10.1186/1471-2105-9-396. BMC Bioinformatics. 2008. PMID: 18816385 Free PMC article.
-
FAF-Drugs3: a web server for compound property calculation and chemical library design.Nucleic Acids Res. 2015 Jul 1;43(W1):W200-7. doi: 10.1093/nar/gkv353. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883137 Free PMC article.
-
The FAF-Drugs2 server: a multistep engine to prepare electronic chemical compound collections.Bioinformatics. 2011 Jul 15;27(14):2018-20. doi: 10.1093/bioinformatics/btr333. Epub 2011 Jun 2. Bioinformatics. 2011. PMID: 21636592
-
In silico ADME-Tox modeling: progress and prospects.Expert Opin Drug Metab Toxicol. 2017 Nov;13(11):1147-1158. doi: 10.1080/17425255.2017.1389897. Epub 2017 Oct 13. Expert Opin Drug Metab Toxicol. 2017. PMID: 28988506 Review.
-
Applying machine learning techniques for ADME-Tox prediction: a review.Expert Opin Drug Metab Toxicol. 2015 Feb;11(2):259-71. doi: 10.1517/17425255.2015.980814. Epub 2014 Dec 2. Expert Opin Drug Metab Toxicol. 2015. PMID: 25440524 Review.
Cited by
-
Applications of Virtual Screening in Bioprospecting: Facts, Shifts, and Perspectives to Explore the Chemo-Structural Diversity of Natural Products.Front Chem. 2021 Apr 29;9:662688. doi: 10.3389/fchem.2021.662688. eCollection 2021. Front Chem. 2021. PMID: 33996755 Free PMC article. Review.
-
Advances in computationally modeling human oral bioavailability.Adv Drug Deliv Rev. 2015 Jun 23;86:11-6. doi: 10.1016/j.addr.2015.01.001. Epub 2015 Jan 9. Adv Drug Deliv Rev. 2015. PMID: 25582307 Free PMC article. Review.
-
Frog: a FRee Online druG 3D conformation generator.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W568-72. doi: 10.1093/nar/gkm289. Epub 2007 May 7. Nucleic Acids Res. 2007. PMID: 17485475 Free PMC article.
-
High Impact: The Role of Promiscuous Binding Sites in Polypharmacology.Molecules. 2019 Jul 10;24(14):2529. doi: 10.3390/molecules24142529. Molecules. 2019. PMID: 31295958 Free PMC article.
-
Identification of new potential Mycobacterium tuberculosis shikimate kinase inhibitors through molecular docking simulations.J Mol Model. 2012 Feb;18(2):755-64. doi: 10.1007/s00894-011-1113-5. Epub 2011 May 19. J Mol Model. 2012. PMID: 21594693
References
-
- Lyne P.D. Structure-based virtual screening: an overview. Drug Discov. Today. 2002;7:1047–1055. - PubMed
-
- Bleicher K.H., Bohm H.J., Muller K., Alanine A.I. Hit and lead generation: beyond high-throughput screening. Nature Rev. Drug Discov. 2003;2:369–378. - PubMed
-
- Jennings A., Tennant M. Discovery strategies in a BioPharmaceutical startup: maximising your chances of success using computational filters. Curr. Pharm. Des. 2005;11:335–344. - PubMed
-
- McConkey B.J., Sobolev V., Edelman M. The performance of current methods in ligand-protein docking. Current Science. 2002;83:845–856.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

