HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary
- PMID: 16845368
- PMCID: PMC1525151
- DOI: 10.1038/sj.embor.7400758
HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary
Abstract
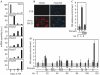
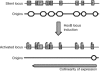
In multicellular organisms, changes in the DNA replication programme could act to integrate differentiation with cell division in various developmental and transcriptional contexts. Here, we have addressed the use of DNA replication origins during differentiation in the HoxB domain-a cluster of nine genes developmentally regulated in a collinear manner. In undifferentiated mouse P19 cells, we detected several DNA replication origins in the 100 kb HoxB locus, indicating a relaxed origin use when the locus is transcriptionally silent. By contrast, in retinoic-acid-induced differentiated cells, when HoxB transcription is activated, a general silencing of DNA replication origins occurs in the locus except one located downstream of Hoxb1, at the 3' boundary of the HoxB domain. Silencing of the replication origins is associated with histone hyperacetylation, whereas the active Hoxb1 origin persists as a hypoacetylated island. These findings provide direct evidence for the differentiated use of origins in HoxB genes, and we suggest that this regulation might contribute to the regulated expression of HoxB genes during development.
Figures

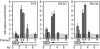

Comment in
-
DNA replication: the unbearable lightness of origins.EMBO Rep. 2006 Aug;7(8):779-81. doi: 10.1038/sj.embor.7400766. EMBO Rep. 2006. PMID: 16880822 Free PMC article. Review. No abstract available.
References
-
- Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 - PubMed
-
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Méchali M (2004) Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6: 721–730 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

