Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout
- PMID: 16845372
- PMCID: PMC1525150
- DOI: 10.1038/sj.embor.7400751
Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout
Abstract
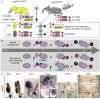
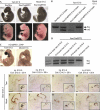
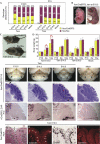
To identify the independent spatial and temporal activities of the essential developmental gene the Otx2, the germline mutation of which is lethal at embryonic day 8.5, we floxed one allele and substituted the other with an inducible CreER recombinase gene. This makes 'trans' self-knockout possible at any developmental stage. The transient action of tamoxifen pulses allows time-course mutation. We demonstrate efficient temporal knockout and demarcate spatio-temporal windows in which Otx2 controls the head, brain structures and body development.
Figures

Similar articles
-
A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors.BMC Dev Biol. 2007 Nov 2;7:122. doi: 10.1186/1471-213X-7-122. BMC Dev Biol. 2007. PMID: 17980036 Free PMC article.
-
Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain.Int J Dev Biol. 2010;54(5):939-45. doi: 10.1387/ijdb.092974ms. Int J Dev Biol. 2010. PMID: 19924631
-
Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage.Genesis. 2009 Dec;47(12):805-14. doi: 10.1002/dvg.20564. Genesis. 2009. PMID: 19830818 Free PMC article.
-
Brain development is a multi-level regulated process--the case of the OTX2 gene.Pediatr Endocrinol Rev. 2011 Sep;9(1):422-30. Pediatr Endocrinol Rev. 2011. PMID: 22783640 Review.
-
The homeobox gene Otx2 in development and disease.Exp Eye Res. 2013 Jun;111:9-16. doi: 10.1016/j.exer.2013.03.007. Epub 2013 Mar 21. Exp Eye Res. 2013. PMID: 23523800 Review.
Cited by
-
A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors.BMC Dev Biol. 2007 Nov 2;7:122. doi: 10.1186/1471-213X-7-122. BMC Dev Biol. 2007. PMID: 17980036 Free PMC article.
-
Animal Models in Psychiatric Disease: A Circuit-Search Approach.Harv Rev Psychiatry. 2018 Sep/Oct;26(5):298-303. doi: 10.1097/HRP.0000000000000193. Harv Rev Psychiatry. 2018. PMID: 30188341 Free PMC article. Review.
-
Aberrant Otx2 expression enhances migration and induces ectopic proliferation of hindbrain neuronal progenitor cells.PLoS One. 2012;7(4):e36211. doi: 10.1371/journal.pone.0036211. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558385 Free PMC article.
-
Haploinsufficiency of Homeodomain Proteins Six3, Vax1, and Otx2 Causes Subfertility in Mice via Distinct Mechanisms.Neuroendocrinology. 2019;109(3):200-207. doi: 10.1159/000494086. Epub 2018 Sep 27. Neuroendocrinology. 2019. PMID: 30261489 Free PMC article. Review.
-
Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism.Mol Endocrinol. 2011 May;25(5):833-46. doi: 10.1210/me.2010-0271. Epub 2011 Mar 24. Mol Endocrinol. 2011. PMID: 21436260 Free PMC article.
References
-
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P (1995) Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121: 3279–3290 - PubMed
-
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J (1996) A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122: 243–252 - PubMed
-
- Di C et al. (2005) Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res 65: 919–924 - PubMed
-
- Feil R, Wagner J, Metzger D, Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237: 752–757 - PubMed
-
- Foucher I, Mione M, Simeone A, Acampora D, Bally-Cuif L, Houart C (2006) Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development 133: 1891–1900 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

