Initial transcribed sequence mutations specifically affect promoter escape properties
- PMID: 16846227
- PMCID: PMC2528282
- DOI: 10.1021/bi060247u
Initial transcribed sequence mutations specifically affect promoter escape properties
Abstract
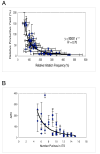
Promoter escape efficiency of E. coli RNA polymerase is guided by both the core promoter and the initial transcribed sequence (ITS). Here, we quantitatively examined the escape properties of 43 random initial sequence variants of the phage T5 N25 promoter. The position for promoter escape on all N25-ITS variants occurred at the +15/+16 juncture, unlike the +11/+12 juncture for the wild type N25. These variants further exhibited a 25-fold difference in escape efficiency. ITS changes favoring promoter escape showed a compositional bias that is unrelated to nucleotide substrate binding affinity for the initial positions. Comparing all variants, the natural N25 promoter emerges as having evolved an ITS optimal for promoter escape, giving a high level of productive synthesis after undergoing the shortest abortive program. We supplemented GreB to transcription reactions to better understand abortive initiation and promoter escape in vivo. GreB supplementation elevated productive RNA synthesis 2-5-fold by altering the abortive RNA pattern, decreasing the abundance of the medium (6-10 nt) to long (11-15 nt) abortive RNAs without changing the levels of short (2-5 nt) and very long abortive RNAs (16-20 nt). The GreB-refractive nature of short abortive RNA production may reflect a minimum length requirement of 4-5 bp of the RNA-DNA hybrid for maintaining the stability of initial or backtracked complexes. That the very long abortive RNAs are unaffected by GreB suggests that they are unlikely to be products of polymerase backtracking. How the ITS might influence the course of early transcription is discussed within the structural context of an initial transcribing complex.
Figures







References
-
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. - PubMed
-
- Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. - PubMed
-
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. - PubMed
-
- Tahirov TH, Temiakov D, Patlan AM, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420:43–50. - PubMed
-
- Murakami KS, Masuda S, Campbell EA, Mussin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

