Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients
- PMID: 16846532
- PMCID: PMC1779468
- DOI: 10.1186/bcr1517
Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients
Abstract
Background: Molecular markers and the rich biological information they contain have great potential for cancer diagnosis, prognostication and therapy prediction. So far, however, they have not superseded routine histopathology and staging criteria, partly because the few studies performed on molecular subtyping have had little validation and limited clinical characterization.
Methods: We obtained gene expression and clinical data for 412 breast cancers obtained from population-based cohorts of patients from Stockholm and Uppsala, Sweden. Using the intrinsic set of approximately 500 genes derived in the Norway/Stanford breast cancer data, we validated the existence of five molecular subtypes--basal-like, ERBB2, luminal A/B and normal-like--and characterized these subtypes extensively with the use of conventional clinical variables.
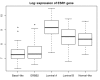
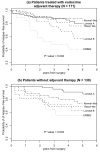
Results: We found an overall 77.5% concordance between the centroid prediction of the Swedish cohort by using the Norway/Stanford signature and the k-means clustering performed internally within the Swedish cohort. The highest rate of discordant assignments occurred between the luminal A and luminal B subtypes and between the luminal B and ERBB2 subtypes. The subtypes varied significantly in terms of grade (p < 0.001), p53 mutation (p < 0.001) and genomic instability (p = 0.01), but surprisingly there was little difference in lymph-node metastasis (p = 0.31). Furthermore, current users of hormone-replacement therapy were strikingly over-represented in the normal-like subgroup (p < 0.001). Separate analyses of the patients who received endocrine therapy and those who did not receive any adjuvant therapy supported the previous hypothesis that the basal-like subtype responded to adjuvant treatment, whereas the ERBB2 and luminal B subtypes were poor responders.
Conclusion: We found that the intrinsic molecular subtypes of breast cancer are broadly present in a diverse collection of patients from a population-based cohort in Sweden. The intrinsic gene set, originally selected to reveal stable tumor characteristics, was shown to have a strong correlation with progression-related properties such as grade, p53 mutation and genomic instability.
Figures
References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. - DOI - PMC - PubMed
-
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. - DOI - PMC - PubMed
-
- Egyhazi S, Bjohle J, Skoog L, Huang F, Borg AL, Frostvik Stolt M, Hagerstrom T, Ringborg U, Bergh J. Proteinase K added to the extraction procedure markedly increases RNA yield from primary breast tumors for use in microarray studies. Clin Chem. 2004;50:975–976. doi: 10.1373/clinchem.2003.027102. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

