Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions
- PMID: 16847312
- PMCID: PMC1592780
- DOI: 10.1128/MCB.00405-06
Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions
Abstract
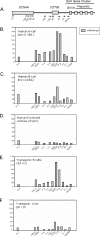
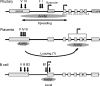
Random assortment within mammalian genomes juxtaposes genes with distinct expression profiles. This organization, along with the prevalence of long-range regulatory controls, generates a potential for aberrant transcriptional interactions. The human CD79b/GH locus contains six tightly linked genes with three mutually exclusive tissue specificities and interdigitated control elements. One consequence of this compact organization is that the pituitary cell-specific transcriptional events that activate hGH-N also trigger ectopic activation of CD79b. However, the B-cell-specific events that activate CD79b do not trigger reciprocal activation of hGH-N. Here we utilized DNase I hypersensitive site mapping, chromatin immunoprecipitation, and transgenic models to explore the basis for this asymmetric relationship. The results reveal tissue-specific patterns of chromatin structures and transcriptional controls at the CD79b/GH locus in B cells distinct from those in the pituitary gland and placenta. These three unique transcriptional environments suggest a set of corresponding gene expression pathways and transcriptional interactions that are likely to be found juxtaposed at multiple sites within the eukaryotic genome.
Figures





Similar articles
-
Locus control region transcription plays an active role in long-range gene activation.Mol Cell. 2006 Aug 4;23(3):365-75. doi: 10.1016/j.molcel.2006.05.041. Mol Cell. 2006. PMID: 16885026
-
Bystander gene activation by a locus control region.EMBO J. 2004 Oct 1;23(19):3854-63. doi: 10.1038/sj.emboj.7600365. Epub 2004 Sep 9. EMBO J. 2004. PMID: 15359275 Free PMC article.
-
Expression of Pit-1 in nonsomatotrope cell lines induces human growth hormone locus control region histone modification and hGH-N transcription.J Mol Biol. 2009 Jul 3;390(1):26-44. doi: 10.1016/j.jmb.2009.04.081. Epub 2009 May 7. J Mol Biol. 2009. PMID: 19427323
-
Activation of the human GH gene cluster: roles for targeted chromatin modification.Trends Endocrinol Metab. 2004 Jan-Feb;15(1):40-5. doi: 10.1016/j.tem.2003.11.004. Trends Endocrinol Metab. 2004. PMID: 14693425 Review.
-
Chromatin looping and the probability of transcription.Trends Genet. 2006 Apr;22(4):197-202. doi: 10.1016/j.tig.2006.02.004. Epub 2006 Feb 21. Trends Genet. 2006. PMID: 16494964 Review.
Cited by
-
Human metastatic melanoma cell lines express high levels of growth hormone receptor and respond to GH treatment.Biochem Biophys Res Commun. 2013 Nov 8;441(1):144-50. doi: 10.1016/j.bbrc.2013.10.023. Epub 2013 Oct 14. Biochem Biophys Res Commun. 2013. PMID: 24134847 Free PMC article.
-
Long-range looping of a locus control region drives tissue-specific chromatin packing within a multigene cluster.Nucleic Acids Res. 2016 Jun 2;44(10):4651-64. doi: 10.1093/nar/gkw090. Epub 2016 Feb 17. Nucleic Acids Res. 2016. PMID: 26893355 Free PMC article.
-
Identification of a secondary promoter within the human B cell receptor component gene hCD79b.J Biol Chem. 2013 Jun 21;288(25):18353-65. doi: 10.1074/jbc.M113.461988. Epub 2013 May 6. J Biol Chem. 2013. PMID: 23649625 Free PMC article.
-
Autonomous actions of the human growth hormone long-range enhancer.Nucleic Acids Res. 2015 Feb 27;43(4):2091-101. doi: 10.1093/nar/gkv093. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662214 Free PMC article.
-
DNase I hypersensitive site II of the human growth hormone locus control region mediates an essential and distinct long-range enhancer function.J Biol Chem. 2012 Jul 20;287(30):25454-65. doi: 10.1074/jbc.M112.365825. Epub 2012 Jun 5. J Biol Chem. 2012. PMID: 22669946 Free PMC article.
References
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 1997. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
-
- Bennani-Baiti, I. M., N. E. Cooke, and S. A. Liebhaber. 1998. Physical linkage of the human growth hormone gene cluster and the CD79b(Igβ/B29) gene. Genomics 48:258-264. - PubMed
-
- Benschop, R. J., and J. C. Cambier. 1999. B cell development: signal transduction by antigen receptors and their surrogates. Curr. Opin. Immunol. 11:143-151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
