Mechanism of action of a distal NF-kappaB-dependent enhancer
- PMID: 16847329
- PMCID: PMC1592769
- DOI: 10.1128/MCB.00271-06
Mechanism of action of a distal NF-kappaB-dependent enhancer
Abstract
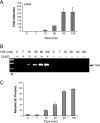
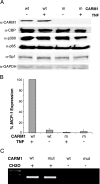
The monocyte chemoattractant protein 1 gene (MCP-1) is regulated by TNF through an NF-kappaB-dependent distal enhancer and an Sp1-dependent promoter-proximal regulatory region. In the silent state, only the distal regulatory region is accessible to transcription factors. Upon activation by tumor necrosis factor, NF-kappaB binds to the distal regulatory region and recruits CBP and p300. CBP and p300 recruitment led to specific histone modifications that ultimately enabled the binding of Sp1 to the proximal regulatory region. During this process, a direct interaction between the distal and proximal regulatory regions occurred. Sp1, NF-kappaB, CBP, and p300 were required for this interaction. CBP/p300-mediated histone modifications enhanced the binding of the coactivator CARM1 to the distal regulatory region. CARM1, which is necessary for MCP-1 expression, was not required for distal-proximal region interactions, suggesting that it plays a later downstream activation event. The results describe a model in which the separation of the distal enhancer from the promoter-proximal region allows for two independent chromatin states to exist, preventing inappropriate gene activation at the promoter while at the same time allowing rapid induction through the distal regulatory region.
Figures

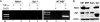


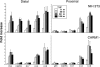
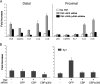
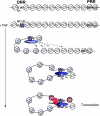
Similar articles
-
Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene.J Immunol. 2003 Apr 15;170(8):4139-47. doi: 10.4049/jimmunol.170.8.4139. J Immunol. 2003. PMID: 12682245
-
Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17.Mol Endocrinol. 2006 Jul;20(7):1562-73. doi: 10.1210/me.2005-0365. Epub 2006 Feb 23. Mol Endocrinol. 2006. PMID: 16497732
-
Nuclear factor-kappa B p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene.J Immunol. 1999 Jan 15;162(2):727-34. J Immunol. 1999. PMID: 9916692
-
Dynamics of sequestering the limiting p300/CBP, viral cis-regulatory elements, and disease.J Biosci. 2020;45:79. J Biosci. 2020. PMID: 32515361 Review.
-
Unique aspects of transcriptional regulation in neurons--nuances in NFkappaB and Sp1-related factors.J Neuroinflammation. 2009 May 18;6:16. doi: 10.1186/1742-2094-6-16. J Neuroinflammation. 2009. PMID: 19450264 Free PMC article. Review.
Cited by
-
CARM1 methylates MED12 to regulate its RNA-binding ability.Life Sci Alliance. 2018 Sep 19;1(5):e201800117. doi: 10.26508/lsa.201800117. eCollection 2018 Oct. Life Sci Alliance. 2018. PMID: 30456381 Free PMC article.
-
CCR5 deficiency accelerates lipopolysaccharide-induced astrogliosis, amyloid-beta deposit and impaired memory function.Oncotarget. 2016 Mar 15;7(11):11984-99. doi: 10.18632/oncotarget.7453. Oncotarget. 2016. PMID: 26910914 Free PMC article.
-
Co-Inflammatory Roles of TGFβ1 in the Presence of TNFα Drive a Pro-inflammatory Fate in Mesenchymal Stem Cells.Front Immunol. 2017 May 11;8:479. doi: 10.3389/fimmu.2017.00479. eCollection 2017. Front Immunol. 2017. PMID: 28553282 Free PMC article.
-
Transcriptional control of maladaptive and protective responses in alcoholics: a role of the NF-κB system.Brain Behav Immun. 2011 Jun;25 Suppl 1(Suppl 1):S29-38. doi: 10.1016/j.bbi.2010.12.019. Epub 2010 Dec 30. Brain Behav Immun. 2011. PMID: 21195164 Free PMC article. Review.
-
IFN-γ and LPS Induce Synergistic Expression of CCL2 in Monocytic Cells via H3K27 Acetylation.J Inflamm Res. 2022 Jul 27;15:4291-4302. doi: 10.2147/JIR.S368352. eCollection 2022. J Inflamm Res. 2022. PMID: 35923906 Free PMC article.
References
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. - PubMed
-
- Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. - PubMed
-
- Baeuerle, P. A., and D. Baltimore. 1996. NF-kappa B: ten years after. Cell 87:13-20. - PubMed
-
- Baggiolini, M., and P. Loetscher. 2000. Chemokines in inflammation and immunity. Immunol. Today 21:418-420. - PubMed
-
- Baldwin, A. S. 1996. The NF-kB and IkB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
