Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain
- PMID: 16847350
- PMCID: PMC1522084
- DOI: 10.1101/gad.1413706
Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain
Abstract
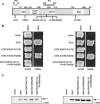
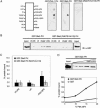
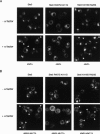
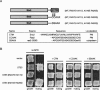
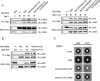
Ste5, the prototypic mitogen-activated protein kinase (MAPK) scaffold protein, associates with plasma membrane-tethered Gbetagamma freed upon pheromone receptor occupancy, thereby initiating downstream signaling. We demonstrate that this interaction and membrane binding of an N-terminal amphipathic alpha-helix (PM motif) are not sufficient for Ste5 action. Rather, Ste5 contains a pleckstrin-homology (PH) domain (residues 388-518) that is essential for its membrane recruitment and function. Altering residues (R407S K411S) equivalent to those that mediate phosphoinositide binding in other PH domains abolishes Ste5 function. The isolated PH domain, but not a R407S K411S derivative, binds phosphoinositides in vitro. Ste5(R407S K411S) is expressed normally, retains Gbetagamma and Ste11 binding, and oligomerizes, yet is not recruited to the membrane in response to pheromone. Artificial membrane tethering of Ste5(R407S K411S) restores signaling. R407S K411S loss-of-function mutations abrogate the constitutive activity of gain-of-function Ste5 alleles, including one (P44L) that increases membrane affinity of the PM motif. Thus, the PH domain is essential for stable membrane recruitment of Ste5, and this association is critical for initiation of downstream signaling because it allows Ste5-bound Ste11 (MAPKKK) to be activated by membrane-bound Ste20 (MAPKKKK).
Figures


Similar articles
-
Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein.Mol Biol Cell. 2000 Nov;11(11):4033-49. doi: 10.1091/mbc.11.11.4033. Mol Biol Cell. 2000. PMID: 11071925 Free PMC article.
-
The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae.Mol Cell Biol. 2006 Feb;26(3):912-28. doi: 10.1128/MCB.26.3.912-928.2006. Mol Cell Biol. 2006. PMID: 16428446 Free PMC article.
-
Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway.Genes Dev. 1998 Sep 1;12(17):2684-97. doi: 10.1101/gad.12.17.2684. Genes Dev. 1998. PMID: 9732267 Free PMC article.
-
Pheromone response, mating and cell biology.Curr Opin Microbiol. 2000 Dec;3(6):573-81. doi: 10.1016/s1369-5274(00)00143-0. Curr Opin Microbiol. 2000. PMID: 11121776 Review.
-
The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast.Curr Genet. 2003 Jun;43(3):161-70. doi: 10.1007/s00294-003-0383-6. Epub 2003 Mar 11. Curr Genet. 2003. PMID: 12764668 Review.
Cited by
-
Localized Surface Plasmon Resonance Biosensing: Current Challenges and Approaches.Sensors (Basel). 2015 Jul 2;15(7):15684-716. doi: 10.3390/s150715684. Sensors (Basel). 2015. PMID: 26147727 Free PMC article. Review.
-
The putative lipid transporter, Arv1, is required for activating pheromone-induced MAP kinase signaling in Saccharomyces cerevisiae.Genetics. 2011 Feb;187(2):455-65. doi: 10.1534/genetics.110.120725. Epub 2010 Nov 23. Genetics. 2011. PMID: 21098723 Free PMC article.
-
Heterotrimeric G Protein-coupled Receptor Signaling in Yeast Mating Pheromone Response.J Biol Chem. 2016 Apr 8;291(15):7788-95. doi: 10.1074/jbc.R116.714980. Epub 2016 Feb 23. J Biol Chem. 2016. PMID: 26907689 Free PMC article. Review.
-
The regulation of filamentous growth in yeast.Genetics. 2012 Jan;190(1):23-49. doi: 10.1534/genetics.111.127456. Genetics. 2012. PMID: 22219507 Free PMC article. Review.
-
Scaffold proteins: hubs for controlling the flow of cellular information.Science. 2011 May 6;332(6030):680-6. doi: 10.1126/science.1198701. Science. 2011. PMID: 21551057 Free PMC article. Review.
References
-
- Butty A.C., Pryciak P.M., Huang L.S., Herskowitz I., Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. - PubMed
-
- Choi K.Y., Satterberg B., Lyons D.M., Elion E.A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. - PubMed
-
- Cole G.M., Reed S.I. Pheromone-induced phosphorylation of a G protein β subunit in S. cerevisiae is associated with an adaptive response to mating pheromone. Cell. 1991;64:703–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
