SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae
- PMID: 16847351
- PMCID: PMC1536058
- DOI: 10.1101/gad.1430406
SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae
Abstract
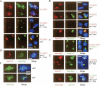
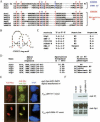
The synaptonemal complex (SC) is a proteinaceous complex that apparently mediates synapsis between homologous chromosomes during meiotic prophase. In Saccharomyces cerevisiae, the Zip1 protein is the integral component of the SC. In the absence of a DNA double-strand break or the SC initiation protein Zip3, Zip1 proteins aggregate to form a polycomplex (PC). In addition, Zip1 is also responsible for DSB-independent nonhomologous centromere coupling at early meiotic prophase. We report here that Zip3 is a SUMO (small ubiquitin-related modifier) E3 ligase and that Zip1 is a binding protein for SUMO-conjugated products. Our results also suggest that at early meiotic prophase, Zip1 interacts with Zip3-independent Smt3 conjugates (e.g., Top2) to promote nonhomologous centromere coupling. At and after mid-prophase, the Zip1 protein begins to associate with Zip3-dependent Smt3 conjugates (e.g., Red1) along meiotic chromosomes in the wild-type cell to form SCs and with Smt3 polymeric chains in the zip3 mutant to form PCs.
Figures



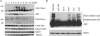

References
-
- Agarwal S., Roeder G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. - PubMed
-
- Aguilar C., Davidson C., Dix M., Stead K., Zheng K., Hartman T., Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. - PubMed
-
- Arora C., Kee K., Maleki S., Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell. 2004;13:549–559. - PubMed
-
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S.J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
