Toward a census of bacteria in soil
- PMID: 16848637
- PMCID: PMC1513271
- DOI: 10.1371/journal.pcbi.0020092
Toward a census of bacteria in soil
Abstract
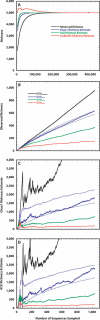
For more than a century, microbiologists have sought to determine the species richness of bacteria in soil, but the extreme complexity and unknown structure of soil microbial communities have obscured the answer. We developed a statistical model that makes the problem of estimating richness statistically accessible by evaluating the characteristics of samples drawn from simulated communities with parametric community distributions. We identified simulated communities with rank-abundance distributions that followed a truncated lognormal distribution whose samples resembled the structure of 16S rRNA gene sequence collections made using Alaskan and Minnesotan soils. The simulated communities constructed based on the distribution of 16S rRNA gene sequences sampled from the Alaskan and Minnesotan soils had a richness of 5,000 and 2,000 operational taxonomic units (OTUs), respectively, where an OTU represents a collection of sequences not more than 3% distant from each other. To sample each of these OTUs in the Alaskan 16S rRNA gene library at least twice, 480,000 sequences would be required; however, to estimate the richness of the simulated communities using nonparametric richness estimators would require only 18,000 sequences. Quantifying the richness of complex environments such as soil is an important step in building an ecological framework. We have shown that generating sufficient sequence data to do so requires less sequencing effort than completely sequencing a bacterial genome.
Conflict of interest statement
Figures




References
-
- Begon M, Harper JL, Townsend CR. Ecology: Individuals, populations, and communities. 3rd ed. Malden (Massachusetts): Blackwell Science; 1996. pp. 829–830.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

