Tempo and mode of early gene loss in endosymbiotic bacteria from insects
- PMID: 16848891
- PMCID: PMC1544356
- DOI: 10.1186/1471-2148-6-56
Tempo and mode of early gene loss in endosymbiotic bacteria from insects
Abstract
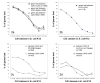
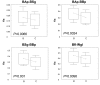
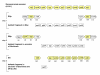
Background: Understanding evolutionary processes that drive genome reduction requires determining the tempo (rate) and the mode (size and types of deletions) of gene losses. In this study, we analysed five endosymbiotic genome sequences of the gamma-proteobacteria (three different Buchnera aphidicola strains, Wigglesworthia glossinidia, Blochmannia floridanus) to test if gene loss could be driven by the selective importance of genes. We used a parsimony method to reconstruct a minimal ancestral genome of insect endosymbionts and quantified gene loss along the branches of the phylogenetic tree. To evaluate the selective or functional importance of genes, we used a parameter that measures the level of adaptive codon bias in E. coli (i.e. codon adaptive index, or CAI), and also estimates of evolutionary rates (Ka) between pairs of orthologs either in free-living bacteria or in pairs of symbionts.
Results: Our results demonstrate that genes lost in the early stages of symbiosis were on average less selectively constrained than genes conserved in any of the extant symbiotic strains studied. These results also extend to more recent events of gene losses (i.e. among Buchnera strains) that still tend to concentrate on genes with low adaptive bias in E. coli and high evolutionary rates both in free-living and in symbiotic lineages. In addition, we analyzed the physical organization of gene losses for early steps of symbiosis acquisition under the hypothesis of a common origin of different symbioses. In contrast with previous findings we show that gene losses mostly occurred through loss of rather small blocks and mostly in syntenic regions between at least one of the symbionts and present-day E. coli.
Conclusion: At both ancient and recent stages of symbiosis evolution, gene loss was at least partially influenced by selection, highly conserved genes being retained more readily than lowly conserved genes: although losses might result from drift due to the bottlenecking of endosymbiontic populations, we demonstrated that purifying selection also acted by retaining genes of greater selective importance.
Figures
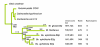


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

