NERVE: new enhanced reverse vaccinology environment
- PMID: 16848907
- PMCID: PMC1570458
- DOI: 10.1186/1472-6750-6-35
NERVE: new enhanced reverse vaccinology environment
Abstract
Background: Since a milestone work on Neisseria meningitidis B, Reverse Vaccinology has strongly enhanced the identification of vaccine candidates by replacing several experimental tasks using in silico prediction steps. These steps have allowed scientists to face the selection of antigens from the predicted proteome of pathogens, for which cell culture is difficult or impossible, saving time and money. However, this good example of bioinformatics-driven immunology can be further developed by improving in silico steps and implementing biologist-friendly tools.
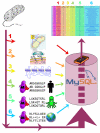
Results: We introduce NERVE (New Enhanced Reverse Vaccinology Environment), an user-friendly software environment for the in silico identification of the best vaccine candidates from whole proteomes of bacterial pathogens. The software integrates multiple robust and well-known algorithms for protein analysis and comparison. Vaccine candidates are ranked and presented in a html table showing relevant information and links to corresponding primary data. Information concerning all proteins of the analyzed proteome is not deleted along selection steps but rather flows into an SQL database for further mining and analyses.
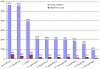
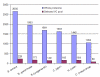
Conclusion: After learning from recent years' works in this field and analysing a large dataset, NERVE has been implemented and tuned as the first available tool able to rank a restricted pool (approximately 8-9% of the whole proteome) of vaccine candidates and to show high recall (approximately 75-80%) of known protective antigens. These vaccine candidates are required to be "safe" (taking into account autoimmunity risk) and "easy" for further experimental, high-throughput screening (avoiding possibly not soluble antigens). NERVE is expected to help save time and money in vaccine design and is available as an additional file with this manuscript; updated versions will be available at http://www.bio.unipd.it/molbinfo.
Figures

References
-
- Montigiani S, Falugi F, Scarselli M, Finco O, Petracca R, Galli G, Mariani M, Manetti R, Agnusdei M, Cevenini R, Donati M, Nogarotto R, Norais N, Garaguso I, Nuti S, Saletti G, Rosa D, Ratti G, Grandi G. Genomic Approach for Analysis of Surface Proteins in Chlamydia pneumoniae. Infect Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. - DOI - PMC - PubMed
-
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JV, Moxon ER, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. - DOI - PubMed
-
- Ross BC, Czajkowski L, Hocking D, Margetts M, Webb E, Rothel L, Patterson M, Agius C, Camuglia S, Reynolds E, Littlejohn T, Gaeta B, Ng A, Kuczek ES, Mattick JS, Gearing D, Barr IG. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19:4135–4142. doi: 10.1016/S0264-410X(01)00173-6. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

