Stochastic models of receptor oligomerization by bivalent ligand
- PMID: 16849251
- PMCID: PMC1664659
- DOI: 10.1098/rsif.2006.0116
Stochastic models of receptor oligomerization by bivalent ligand
Abstract
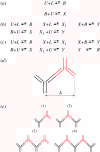
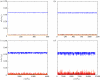
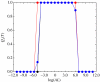
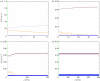
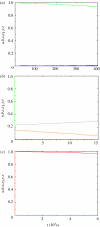
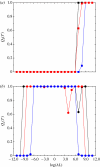
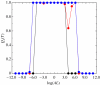
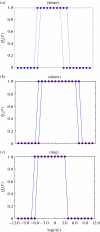
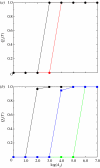
In this paper, we develop stochastic models of receptor binding by a bivalent ligand. A detailed kinetic study allows us to analyse the role of cross-linking in cell activation by receptor oligomerization. We show how oligomer formation could act to buffer intracellular signalling against stochastic fluctuations. In addition, we put forward the hypothesis that formation of long linear oligomers increases the range of ligand concentration to which the cell is responsive, whereas formation of closed oligomers increases ligand concentration specificity. Thus, different physiological functions requiring different degrees of specificity to ligand concentration would favour formation of oligomers with different lengths and geometries. Furthermore, provided that ligand concentration specificity is taken as a design principle, our model enables us to estimate parameters, such as the minimum proportion of receptors, that must engage in oligomer formation in order to trigger a cellular response.
Figures



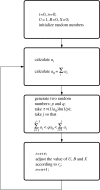
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Garland; New York: 2002. Molecular biology of the cell.
-
- Banu Y, Watanabe T. Augmentation of antigen receptor-mediated responses by histamine H1 receptor signalling. J. Exp. Med. 1999;189:673–682. doi:10.1084/jem.189.4.673 - DOI - PMC - PubMed
-
- Burrage K, Tian T, Burrage P. A multi-scaled approach for simulating chemical reaction systems. Prog. Biophys. Mol. Biol. 2004;85:217–234. doi:10.1016/j.pbiomolbio.2004.01.014 - DOI - PubMed
-
- Durrett R, Levin S. The importance of being discrete (and spatial) Theor. Popul. Biol. 1994;46:363–394. doi:10.1006/tpbi.1994.1032 - DOI
-
- Gardiner C.W. Springer; Berlin: 1983. Handbook of stochastic methods.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

