Globalization and the population structure of Toxoplasma gondii
- PMID: 16849431
- PMCID: PMC1544101
- DOI: 10.1073/pnas.0601438103
Globalization and the population structure of Toxoplasma gondii
Abstract
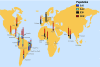
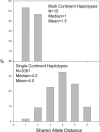
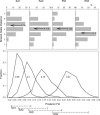
Toxoplasma gondii is a protozoan parasite that infects nearly all mammal and bird species worldwide. Usually asymptomatic, toxoplasmosis can be severe and even fatal to many hosts, including people. Elucidating the contribution of genetic variation among parasites to patterns of disease transmission and manifestations has been the goal of many studies. Focusing on the geographic component of this variation, we show that most genotypes are locale-specific, but some are found across continents and are closely related to each other, indicating a recent radiation of a pandemic genotype. Furthermore, we show that the geographic structure of T. gondii is extraordinary in having one population that is found in all continents except South America, whereas other populations are generally confined to South America, and yet another population is found worldwide. Our evidence suggests that South American and Eurasian populations have evolved separately until recently, when ships populated by rats, mice, and cats provided T. gondii with unprecedented migration opportunities, probably during the transatlantic slave trade. Our results explain several enigmatic features of the population structure of T. gondii and demonstrate how pervasive, prompt, and elusive the impact of human globalization is on nature.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Lindsay D. S., Blagburn B. L., Dubey J. P. Vet. Parasitol. 2002;103:309–313. - PubMed
-
- Dubey J. P. J. Parasitol. 1998;84:862–865. - PubMed
-
- Dubey J. P., Beattie C. P. Toxoplasmosis of Animals and Man. Boca Raton, FL: CRC; 1988.
-
- Dardé M. L., Bouteille B., Pestre-Alexandre M. J. Parasitol. 1992;78:786–794. - PubMed
-
- Howe D. K., Sibley L. D. J. Infect. Dis. 1995;172:1561–1566. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

