Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer
- PMID: 16849555
- PMCID: PMC2933218
- DOI: 10.1158/0008-5472.CAN-06-0515
Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer
Abstract
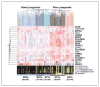
Nonmalignant human mammary epithelial cells (HMEC) seeded in laminin-rich extracellular matrix (lrECM) form polarized acini and, in doing so, transit from a disorganized proliferating state to an organized growth-arrested state. We hypothesized that the gene expression pattern of organized and growth-arrested HMECs would share similarities with breast tumors with good prognoses. Using Affymetrix HG-U133A microarrays, we analyzed the expression of 22,283 gene transcripts in 184 (finite life span) and HMT3522 S1 (immortal nonmalignant) HMECs on successive days after seeding in a lrECM assay. Both HMECs underwent growth arrest in G0-G1 and differentiated into polarized acini between days 5 and 7. We identified gene expression changes with the same temporal pattern in both lines and examined the expression of these genes in a previously published panel of microarray data for 295 breast cancer samples. We show that genes that are significantly lower in the organized, growth-arrested HMEC than in their proliferating counterparts can be used to classify breast cancer patients into poor and good prognosis groups with high accuracy. This study represents a novel unsupervised approach to identifying breast cancer markers that may be of use clinically.
Figures




References
-
- Stampfer MR, Yaswen P. Culture systems for study of human mammary epithelial cell proliferation, differentiation, and transformation. Cancer Surv. 1993;18:7–34. - PubMed
-
- Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–8. - PubMed
-
- Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–92. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

