An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function
- PMID: 16849596
- PMCID: PMC1569793
- DOI: 10.1534/genetics.106.061705
An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function
Abstract
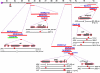
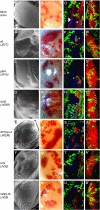
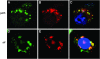
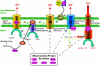
We conducted a screen for glossy-eye flies that fail to incorporate BrdU in the third larval instar eye disc but exhibit normal neuronal differentiation and isolated 23 complementation groups of mutants. These same phenotypes were previously seen in mutants for cytochrome c oxidase subunit Va. We have molecularly characterized six complementation groups and, surprisingly, each encodes a mitochondrial protein. Therefore, we believe our screen to be an efficient method for identifying genes with mitochondrial function.
Figures

Similar articles
-
Mutations in the Drosophila mitochondrial tRNA amidotransferase, bene/gatA, cause growth defects in mitotic and endoreplicating tissues.Genetics. 2008 Feb;178(2):979-87. doi: 10.1534/genetics.107.084376. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245325 Free PMC article.
-
Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae.Genome Biol. 2005;6(2):R11. doi: 10.1186/gb-2005-6-2-r11. Epub 2005 Jan 31. Genome Biol. 2005. PMID: 15693940 Free PMC article.
-
A screen for dominant modifiers of the irreC-rst cell death phenotype in the developing Drosophila retina.Genetics. 2000 Sep;156(1):205-17. doi: 10.1093/genetics/156.1.205. Genetics. 2000. PMID: 10978286 Free PMC article.
-
The art and design of genetic screens: Drosophila melanogaster.Nat Rev Genet. 2002 Mar;3(3):176-88. doi: 10.1038/nrg751. Nat Rev Genet. 2002. PMID: 11972155 Review.
-
The Heidelberg Screen for Pattern Mutants of Drosophila: A Personal Account.Annu Rev Cell Dev Biol. 2016 Oct 6;32:1-46. doi: 10.1146/annurev-cellbio-113015-023138. Epub 2016 Aug 3. Annu Rev Cell Dev Biol. 2016. PMID: 27501451 Review.
Cited by
-
Genetic analysis of fibroblast growth factor signaling in the Drosophila eye.G3 (Bethesda). 2012 Jan;2(1):23-8. doi: 10.1534/g3.111.001495. Epub 2012 Jan 1. G3 (Bethesda). 2012. PMID: 22384378 Free PMC article.
-
A Genetic Screen Using the Drosophila melanogaster TRiP RNAi Collection To Identify Metabolic Enzymes Required for Eye Development.G3 (Bethesda). 2019 Jul 9;9(7):2061-2070. doi: 10.1534/g3.119.400193. G3 (Bethesda). 2019. PMID: 31036678 Free PMC article.
-
Glycolytic disruption restricts Drosophila melanogaster larval growth via the cytokine Upd3.PLoS Genet. 2025 May 2;21(5):e1011690. doi: 10.1371/journal.pgen.1011690. eCollection 2025 May. PLoS Genet. 2025. PMID: 40315265 Free PMC article.
-
Drosophila follicle stem cells are regulated by proliferation and niche adhesion as well as mitochondria and ROS.Nat Commun. 2012 Apr 3;3:769. doi: 10.1038/ncomms1765. Nat Commun. 2012. PMID: 22473013 Free PMC article.
-
Expression profiling of attenuated mitochondrial function identifies retrograde signals in Drosophila.G3 (Bethesda). 2012 Aug;2(8):843-51. doi: 10.1534/g3.112.002584. Epub 2012 Aug 1. G3 (Bethesda). 2012. PMID: 22908033 Free PMC article.
References
-
- Ackerman, S. H., and A. Tzagoloff, 2005. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 80: 95–133. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Ashby, M. N., S. Y. Kutsunai, S. Ackerman, A. Tzagoloff and P. A. Edwards, 1992. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267: 4128–4136. - PubMed
-
- Boyer, P. D., 1997. The ATP synthase: a splendid molecular machine. Annu. Rev. Biochem. 66: 717–749. - PubMed
-
- Claros, M. G., and P. Vincens, 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779–786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

