Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GTPase
- PMID: 16849600
- PMCID: PMC1602101
- DOI: 10.1534/genetics.106.060657
Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GTPase
Abstract
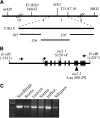
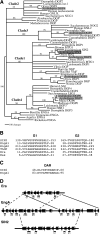
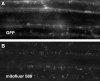
The Arabidopsis short integuments 2-1 (sin2-1) mutant produces ovules with short integuments due to early cessation of cell division in these structures. SIN2 was isolated and encodes a putative GTPase sharing features found in the novel DAR GTPase family. DAR proteins share a signature DAR motif and a unique arrangement of the four conserved GTPase G motifs. We found that DAR GTPases are present in all examined prokaryotes and eukaryotes and that they have diversified into four paralogous lineages in higher eukaryotes. Eukaryotic members of the SIN2 clade of DAR GTPases have been found to localize to mitochondria and are related to eubacterial proteins that facilitate essential steps in biogenesis of the large ribosomal subunit. We propose a similar role for SIN2 in mitochondria. A sin2 insertional allele has ovule effects similar to sin2-1, but more pronounced pleiotropic effects on vegetative and floral development. The diverse developmental effects of the mitochondrial SIN2 GTPase support a mitochondrial role in the regulation of multiple developmental pathways.
Figures

Similar articles
-
A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division.Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5727-31. doi: 10.1073/pnas.082663299. Proc Natl Acad Sci U S A. 2002. PMID: 11960028 Free PMC article.
-
EMB2473/MIRO1, an Arabidopsis Miro GTPase, is required for embryogenesis and influences mitochondrial morphology in pollen.Plant Cell. 2008 Mar;20(3):589-601. doi: 10.1105/tpc.107.055756. Epub 2008 Mar 14. Plant Cell. 2008. PMID: 18344283 Free PMC article.
-
MIRO1 influences the morphology and intracellular distribution of mitochondria during embryonic cell division in Arabidopsis.Plant Cell Rep. 2011 Feb;30(2):239-44. doi: 10.1007/s00299-010-0926-5. Epub 2010 Oct 8. Plant Cell Rep. 2011. PMID: 20931334
-
SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development.Genetics. 2000 Jun;155(2):899-907. doi: 10.1093/genetics/155.2.899. Genetics. 2000. PMID: 10835408 Free PMC article.
-
Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond.J Membr Biol. 1999 May 15;169(2):75-81. doi: 10.1007/s002329900519. J Membr Biol. 1999. PMID: 10341029 Review. No abstract available.
Cited by
-
Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development.Proc Natl Acad Sci U S A. 2011 Mar 29;108(13):5461-5. doi: 10.1073/pnas.1014514108. Epub 2011 Mar 14. Proc Natl Acad Sci U S A. 2011. PMID: 21402944 Free PMC article.
-
Gynoecium size and ovule number are interconnected traits that impact seed yield.J Exp Bot. 2020 May 9;71(9):2479-2489. doi: 10.1093/jxb/eraa050. J Exp Bot. 2020. PMID: 32067041 Free PMC article. Review.
-
Flower development.Arabidopsis Book. 2010;8:e0127. doi: 10.1199/tab.0127. Epub 2010 Mar 23. Arabidopsis Book. 2010. PMID: 22303253 Free PMC article.
-
Mitochondrial biogenesis and function in Arabidopsis.Arabidopsis Book. 2008;6:e0111. doi: 10.1199/tab.0111. Epub 2008 Jul 9. Arabidopsis Book. 2008. PMID: 22303236 Free PMC article.
-
Seed coat thickness in the evolution of angiosperms.Cell Mol Life Sci. 2018 Jul;75(14):2509-2518. doi: 10.1007/s00018-018-2816-x. Epub 2018 May 5. Cell Mol Life Sci. 2018. PMID: 29730767 Free PMC article. Review.
References
-
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski et al., 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8: 517–529. - PubMed
-
- Baum, S. F., and T. L. Rost, 1996. Root apical organization in Arabidopsis thaliana. 1. Root cap and protoderm. Protoplasma 192: 178–188.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

