Regulation of DNA replication machinery by Mrc1 in fission yeast
- PMID: 16849602
- PMCID: PMC1569812
- DOI: 10.1534/genetics.106.060053
Regulation of DNA replication machinery by Mrc1 in fission yeast
Abstract
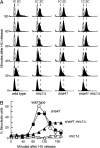
Faithful replication of chromosomes is crucial to genome integrity. In yeast, the ORC binds replication origins throughout the cell cycle. However, Cdc45 binds these before S-phase, and, during replication, it moves along the DNA with MCM helicase. When replication progression is inhibited, checkpoint regulation is believed to stabilize the replication fork; the detailed mechanism, however, remains unclear. To examine the relationship between replication initiation and elongation defects and the response to replication elongation block, we used fission yeast mutants of Orc1 and Cdc45--orp1-4 and sna41-928, respectively--at their respective semipermissive temperatures with regard to BrdU incorporation. Both orp1 and sna41 cells exhibited HU hypersensitivity in the absence of Chk1, a DNA damage checkpoint kinase, and were defective in full activation of Cds1, a replication checkpoint kinase, indicating that normal replication is required for Cds1 activation. Mrc1 is required to activate Cds1 and prevent the replication machinery from uncoupling from DNA synthesis. We observed that, while either the orp1 or the sna41 mutation partially suppressed HU sensitivity of cds1 cells, sna41 specifically suppressed that of mrc1 cells. Interestingly, sna41 alleviated the defect in recovery from HU arrest without increasing Cds1 activity. In addition to sna41, specific mutations of MCM suppressed the HU sensitivity of mrc1 cells. Thus, during elongation, Mrc1 may negatively regulate Cdc45 and MCM helicase to render stalled forks capable of resuming replication.
Figures




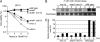
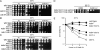
Similar articles
-
Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast.Mol Cell Biol. 2003 Nov;23(22):8395-403. doi: 10.1128/MCB.23.22.8395-8403.2003. Mol Cell Biol. 2003. PMID: 14585996 Free PMC article.
-
Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast.EMBO J. 2007 Mar 7;26(5):1315-26. doi: 10.1038/sj.emboj.7601538. Epub 2007 Feb 15. EMBO J. 2007. PMID: 17304223 Free PMC article.
-
Cds1 phosphorylation by Rad3-Rad26 kinase is mediated by forkhead-associated domain interaction with Mrc1.J Biol Chem. 2004 Jul 30;279(31):32079-86. doi: 10.1074/jbc.M404834200. Epub 2004 Jun 1. J Biol Chem. 2004. PMID: 15173168
-
[Replication fork stabilization by replication stress checkpoint control].Tanpakushitsu Kakusan Koso. 2009 Mar;54(4 Suppl):380-7. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089479 Review. Japanese. No abstract available.
-
Cdc7 kinases (DDKs) and checkpoint responses: lessons from two yeasts.Mutat Res. 2003 Nov 27;532(1-2):21-7. doi: 10.1016/j.mrfmmm.2003.08.007. Mutat Res. 2003. PMID: 14643426 Review.
Cited by
-
Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint.Mol Cell. 2008 Oct 10;32(1):106-17. doi: 10.1016/j.molcel.2008.08.020. Mol Cell. 2008. PMID: 18851837 Free PMC article.
-
Hydroxyurea-The Good, the Bad and the Ugly.Genes (Basel). 2021 Jul 19;12(7):1096. doi: 10.3390/genes12071096. Genes (Basel). 2021. PMID: 34356112 Free PMC article. Review.
-
Replication fork reversal and the maintenance of genome stability.Nucleic Acids Res. 2009 Jun;37(11):3475-92. doi: 10.1093/nar/gkp244. Epub 2009 Apr 30. Nucleic Acids Res. 2009. PMID: 19406929 Free PMC article. Review.
-
Phosphorylation of minichromosome maintenance protein 7 (MCM7) by cyclin/cyclin-dependent kinase affects its function in cell cycle regulation.J Biol Chem. 2013 Jul 5;288(27):19715-25. doi: 10.1074/jbc.M112.449652. Epub 2013 May 17. J Biol Chem. 2013. PMID: 23720738 Free PMC article.
-
Fission yeast Swi1-Swi3 complex facilitates DNA binding of Mrc1.J Biol Chem. 2010 Dec 17;285(51):39609-22. doi: 10.1074/jbc.M110.173344. Epub 2010 Oct 5. J Biol Chem. 2010. PMID: 20924116 Free PMC article.
References
-
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler et al., 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. - PubMed
-
- Alfa, C., P. Fantes, J. Hyams, M. Mcleod and E. Warbrick, 1993. Experiments With Fission Yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Aparicio, O. M., D. M. Weinstein and S. P. Bell, 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69. - PubMed
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. - PubMed
-
- Bell, S. P., and A. Dutta, 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

