MANET: tracing evolution of protein architecture in metabolic networks
- PMID: 16854231
- PMCID: PMC1559654
- DOI: 10.1186/1471-2105-7-351
MANET: tracing evolution of protein architecture in metabolic networks
Abstract
Background: Cellular metabolism can be characterized by networks of enzymatic reactions and transport processes capable of supporting cellular life. Our aim is to find evolutionary patterns and processes embedded in the architecture and function of modern metabolism, using information derived from structural genomics.
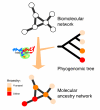
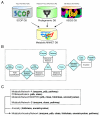
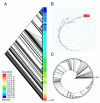
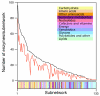
Description: The Molecular Ancestry Network (MANET) project traces evolution of protein architecture in biomolecular networks. We describe metabolic MANET, a database that links information in the Structural Classification of Proteins (SCOP), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and phylogenetic reconstructions depicting the evolution of protein fold architecture. Metabolic MANET literally 'paints' the ancestries of enzymes derived from rooted phylogenomic trees directly onto over one hundred metabolic subnetworks, enabling the study of evolutionary patterns at global and local levels. An initial analysis of painted subnetworks reveals widespread enzymatic recruitment and an early origin of amino acid metabolism.
Conclusion: MANET maps evolutionary relationships directly and globally onto biological networks, and can generate and test hypotheses related to evolution of metabolism. We anticipate its use in the study of other networks, such as signaling and other protein-protein interaction networks.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

