The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda)
- PMID: 16854241
- PMCID: PMC1544340
- DOI: 10.1186/1471-2164-7-182
The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda)
Abstract
Background: Mitochondria contain small genomes that are physically separate from those of nuclei. Their comparison serves as a model system for understanding the processes of genome evolution. Although complete mitochondrial genome sequences have been reported for more than 600 animals, the taxonomic sampling is highly biased toward vertebrates and arthropods, leaving much of the diversity yet uncharacterized.
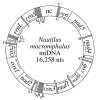
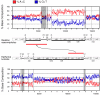
Results: The mitochondrial genome of the bellybutton nautilus, Nautilus macromphalus, a cephalopod mollusk, is 16,258 nts in length and 59.5% A+T, both values that are typical of animal mitochondrial genomes. It contains the 37 genes that are almost universally found in animal mtDNAs, with 15 on one DNA strand and 22 on the other. The arrangement of these genes can be derived from that of the distantly related Katharina tunicata (Mollusca: Polyplacophora) by a switch in position of two large blocks of genes and transpositions of four tRNA genes. There is strong skew in the distribution of nucleotides between the two strands, and analysis of this yields insight into modes of transcription and replication. There is an unusual number of non-coding regions and their function, if any, is not known; however, several of these demark abrupt shifts in nucleotide skew, and there are several identical sequence elements at these junctions, suggesting that they may play roles in transcription and/or replication. One of the non-coding regions contains multiple repeats of a tRNA-like sequence. Some of the tRNA genes appear to overlap on the same strand, but this could be resolved if the polycistron were cleaved at the beginning of the downstream gene, followed by polyadenylation of the product of the upstream gene to form a fully paired structure.
Conclusion: Nautilus macromphalus mtDNA contains an expected gene content that has experienced few rearrangements since the evolutionary split between cephalopods and polyplacophorans. It contains an unusual number of non-coding regions, especially considering that these otherwise often are generated by the same processes that produce gene rearrangements. The skew in nucleotide composition between the two strands is strong and associated with the direction of transcription in various parts of the genomes, but a comparison with K. tunicata implies that mutational bias during replication also plays a role. This appears to be yet another case where polyadenylation of mitochondrial tRNAs restores what would otherwise be an incomplete structure.
Figures


Similar articles
-
The complete mitochondrial genome of the Antarctic springtail Cryptopygus antarcticus (Hexapoda: Collembola).BMC Genomics. 2008 Jul 1;9:315. doi: 10.1186/1471-2164-9-315. BMC Genomics. 2008. PMID: 18593463 Free PMC article.
-
The complete mitochondrial genome of the stomatopod crustacean Squilla mantis.BMC Genomics. 2005 Aug 9;6:105. doi: 10.1186/1471-2164-6-105. BMC Genomics. 2005. PMID: 16091132 Free PMC article.
-
Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis-a case of "Tandem duplication-random loss" for genome rearrangement in Crassostrea?BMC Genomics. 2008 Oct 11;9:477. doi: 10.1186/1471-2164-9-477. BMC Genomics. 2008. PMID: 18847502 Free PMC article.
-
The Drosophila mitochondrial genome.Oxf Surv Eukaryot Genes. 1984;1:1-35. Oxf Surv Eukaryot Genes. 1984. PMID: 6400770 Review.
-
Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system.Gene. 1999 Sep 30;238(1):195-209. doi: 10.1016/s0378-1119(99)00270-x. Gene. 1999. PMID: 10570997 Review.
Cited by
-
A Mitochondrial Genome of Rhyparochromidae (Hemiptera: Heteroptera) and a Comparative Analysis of Related Mitochondrial Genomes.Sci Rep. 2016 Oct 19;6:35175. doi: 10.1038/srep35175. Sci Rep. 2016. PMID: 27756915 Free PMC article.
-
Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications.BMC Genomics. 2015 Jun 16;16(1):460. doi: 10.1186/s12864-015-1679-x. BMC Genomics. 2015. PMID: 26076960 Free PMC article.
-
The complete mitochondrial genome of the stalk-eyed bug Chauliops fallax Scott, and the monophyly of Malcidae (Hemiptera: Heteroptera).PLoS One. 2013;8(2):e55381. doi: 10.1371/journal.pone.0055381. Epub 2013 Feb 4. PLoS One. 2013. PMID: 23390534 Free PMC article.
-
The first mitochondrial genome for the fishfly subfamily Chauliodinae and implications for the higher phylogeny of Megaloptera.PLoS One. 2012;7(10):e47302. doi: 10.1371/journal.pone.0047302. Epub 2012 Oct 9. PLoS One. 2012. PMID: 23056623 Free PMC article.
-
Mitogenomics Reveals a Novel Genetic Code in Hemichordata.Genome Biol Evol. 2019 Jan 1;11(1):29-40. doi: 10.1093/gbe/evy254. Genome Biol Evol. 2019. PMID: 30476024 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

