Organization of the integrin LFA-1 in nanoclusters regulates its activity
- PMID: 16855029
- PMCID: PMC1635357
- DOI: 10.1091/mbc.e05-12-1098
Organization of the integrin LFA-1 in nanoclusters regulates its activity
Abstract
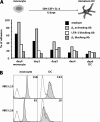
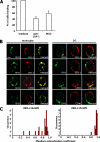
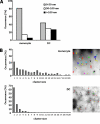
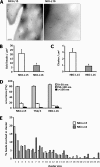
The beta2-integrin LFA-1 facilitates extravasation of monocytes (MOs) into the underlying tissues, where MOs can differentiate into dendritic cells (DCs). Although DCs express LFA-1, unlike MOs, they cannot bind to ICAM-1. We hypothesized that an altered integrin organization on the DC plasma membrane might cause this effect and investigated the relationship between membrane organization and function of LFA-1 on MOs and DCs. High-resolution mapping of LFA-1 surface distribution revealed that on MOs LFA-1 function is associated with a distribution in well-defined nanoclusters (100-150-nm diameter). Interestingly, a fraction of these nanoclusters contains primed LFA-1 molecules expressing the specific activation-dependent L16-epitope. Live imaging of MO-T-cell conjugates showed that only these primed nanoclusters are dynamically recruited to the cellular interface forming micrometer-sized assemblies engaged in ligand binding and linked to talin. We conclude that besides affinity regulation, LFA-1 function is controlled by at least three different avidity patterns: random distributed inactive molecules, well-defined ligand-independent proactive nanoclusters, and ligand-triggered micrometer-sized macroclusters.
Figures




References
-
- Banchereau J., Steinman R. M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. - PubMed
-
- Bazzoni G., Hemler M. E. Are changes in integrin affinity and conformation overemphasized? Trends Biochem. Sci. 1998;23:30–34. - PubMed
-
- Beals C. R., Edwards A. C., Gottschalk R. J., Kuijpers T. W., Staunton D. E. CD18 activation epitopes induced by leukocyte activation. J. Immunol. 2001;167:6113–6122. - PubMed
-
- Beglova N., Blacklow S. C., Takagi J., Springer T. A. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat. Struct. Biol. 2002;9:282–287. - PubMed
-
- Bounou S., Giguere J. F., Cantin R., Gilbert C., Imbeault M., Martin G., Tremblay M. J. The importance of virus-associated host ICAM-1 in human immunodeficiency virus type 1 dissemination depends on the cellular context. FASEB J. 2004;18:1294–1296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

