The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate
- PMID: 16855237
- PMCID: PMC1540016
- DOI: 10.1128/JB.00514-06
The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate
Abstract
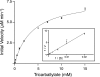
Tricarballylate is the causative agent of grass tetany, a ruminant disease characterized by acute magnesium deficiency. Tricarballylate toxicity has been attributed to its ability to chelate magnesium and to inhibit aconitase, a Krebs cycle enzyme. Neither the ruminant nor the normal rumen flora can catabolize tricarballylate to ameliorate its toxic effects. However, the gram-negative enterobacterium Salmonella enterica can use tricarballylate as a carbon and energy source, providing an opportunity to study the genes and enzymes required for tricarballylate catabolism. The tricarballylate utilization (tcu) genes are organized into two transcriptional units, i.e., tcuR and tcuABC. Here, we report the initial biochemical analysis of TcuA. TcuA catalyzed the oxidation of tricarballylate to cis-aconitate. The apparent K(m) of TcuA for tricarballylate was 3.8 +/- 0.4 mM, with a V(max) of 7.9 +/- 0.3 mM min(-1), turnover number (k(cat)) of 6.7 x 10(-2) s(-1), and a catalytic efficiency (k(cat)/K(m)) of 17.8 M(-1) s(-1). Optimal activity was measured at pH 7.5 and 30 degrees C. The enzyme was inactivated at 45 degrees C. One mole of FAD was present per mole of TcuA. We propose a role for TcuB as an electron shuttle protein responsible for oxidizing FADH(2) back to FAD in TcuA.
Figures
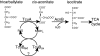





Similar articles
-
Regulation of tricarboxylate transport and metabolism in Acinetobacter baylyi ADP1.Appl Environ Microbiol. 2024 Feb 21;90(2):e0211123. doi: 10.1128/aem.02111-23. Epub 2024 Jan 30. Appl Environ Microbiol. 2024. PMID: 38289138 Free PMC article.
-
Tricarballylate catabolism in Salmonella enterica. The TcuB protein uses 4Fe-4S clusters and heme to transfer electrons from FADH2 in the tricarballylate dehydrogenase (TcuA) enzyme to electron acceptors in the cell membrane.Biochemistry. 2007 Aug 7;46(31):9107-15. doi: 10.1021/bi7006564. Epub 2007 Jul 14. Biochemistry. 2007. PMID: 17630784
-
The Tricarballylate utilization (tcuRABC) genes of Salmonella enterica serovar Typhimurium LT2.J Bacteriol. 2004 Mar;186(6):1629-37. doi: 10.1128/JB.186.6.1629-1637.2004. J Bacteriol. 2004. PMID: 14996793 Free PMC article.
-
Ability of Acidaminococcus fermentans to oxidize trans-aconitate and decrease the accumulation of tricarballylate, a toxic end product of ruminal fermentation.Appl Environ Microbiol. 1994 Jul;60(7):2533-7. doi: 10.1128/aem.60.7.2533-2537.1994. Appl Environ Microbiol. 1994. PMID: 8074529 Free PMC article.
-
Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs.Protein Sci. 1998 Jan;7(1):7-20. doi: 10.1002/pro.5560070102. Protein Sci. 1998. PMID: 9514256 Free PMC article. Review.
Cited by
-
Leveraging a Structural Blueprint to Rationally Engineer the Rieske Oxygenase TsaM.Biochemistry. 2023 Jun 6;62(11):1807-1822. doi: 10.1021/acs.biochem.3c00150. Epub 2023 May 15. Biochemistry. 2023. PMID: 37188334 Free PMC article.
-
Regulation of tricarboxylate transport and metabolism in Acinetobacter baylyi ADP1.Appl Environ Microbiol. 2024 Feb 21;90(2):e0211123. doi: 10.1128/aem.02111-23. Epub 2024 Jan 30. Appl Environ Microbiol. 2024. PMID: 38289138 Free PMC article.
-
Salmonella enterica requires ApbC function for growth on tricarballylate: evidence of functional redundancy between ApbC and IscU.J Bacteriol. 2008 Jul;190(13):4596-602. doi: 10.1128/JB.00262-08. Epub 2008 Apr 25. J Bacteriol. 2008. PMID: 18441067 Free PMC article.
-
Filamentous ascomycetes fungi as a source of natural pigments.Fungal Biol Biotechnol. 2017 May 10;4:4. doi: 10.1186/s40694-017-0033-2. eCollection 2017. Fungal Biol Biotechnol. 2017. PMID: 28955473 Free PMC article. Review.
-
Wavelengths and irradiances modulate the circadian rhythm of Neurospora crassa.PLoS One. 2022 Mar 30;17(3):e0266266. doi: 10.1371/journal.pone.0266266. eCollection 2022. PLoS One. 2022. PMID: 35353854 Free PMC article.
References
-
- Aliverti, A., B. Curti, and M. A. Vanoni. 1999. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins, p. 9-23. In S. K. Chapman and G. A. Reid (ed.), Flavoprotein protocols, vol. 131. Humana Press, Totowa, N.J. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-255. - PubMed
-
- Burau, R., and P. R. Stout. 1965. trans-Aconitic acid in range grasses in early spring. Science 150:766-767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

