Crystal structure of a type III pantothenate kinase: insight into the mechanism of an essential coenzyme A biosynthetic enzyme universally distributed in bacteria
- PMID: 16855243
- PMCID: PMC1540032
- DOI: 10.1128/JB.00469-06
Crystal structure of a type III pantothenate kinase: insight into the mechanism of an essential coenzyme A biosynthetic enzyme universally distributed in bacteria
Abstract
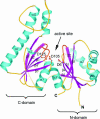
Pantothenate kinase (PanK) catalyzes the first step in the five-step universal pathway of coenzyme A (CoA) biosynthesis, a key transformation that generally also regulates the intracellular concentration of CoA through feedback inhibition. A novel PanK protein encoded by the gene coaX was recently identified that is distinct from the previously characterized type I PanK (exemplified by the Escherichia coli coaA-encoded PanK protein) and type II eukaryotic PanKs and is not inhibited by CoA or its thioesters. This type III PanK, or PanK-III, is widely distributed in the bacterial kingdom and accounts for the only known PanK in many pathogenic species, such as Helicobacter pylori, Bordetella pertussis, and Pseudomonas aeruginosa. Here we report the first crystal structure of a type III PanK, the enzyme from Thermotoga maritima (PanK(Tm)), solved at 2.0-A resolution. The structure of PanK(Tm) reveals that type III PanKs belong to the acetate and sugar kinase/heat shock protein 70/actin (ASKHA) protein superfamily and that they retain the highly conserved active site motifs common to all members of this superfamily. Comparative structural analysis of the PanK(Tm) active site configuration and mutagenesis of three highly conserved active site aspartates identify these residues as critical for PanK-III catalysis. Furthermore, the analysis also provides an explanation for the lack of CoA feedback inhibition by the enzyme. Since PanK-III adopts a different structural fold from that of the E. coli PanK -- which is a member of the "P-loop kinase"superfamily -- this finding represents yet another example of convergent evolution of the same biological function from a different protein ancestor.
Figures
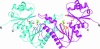

Similar articles
-
Regulation of Coenzyme A Biosynthesis in the Hyperthermophilic Bacterium Thermotoga maritima.J Bacteriol. 2016 Jun 27;198(14):1993-2000. doi: 10.1128/JB.00077-16. Print 2016 Jul 15. J Bacteriol. 2016. PMID: 27161115 Free PMC article.
-
Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.Biochemistry. 2008 Feb 5;47(5):1369-80. doi: 10.1021/bi7018578. Epub 2008 Jan 11. Biochemistry. 2008. PMID: 18186650
-
In Vitro Production of Coenzyme A Using Thermophilic Enzymes.Appl Environ Microbiol. 2021 Jun 25;87(14):e0054121. doi: 10.1128/AEM.00541-21. Epub 2021 Jun 25. Appl Environ Microbiol. 2021. PMID: 33990309 Free PMC article.
-
Physiological roles of the pantothenate kinases.Biochem Soc Trans. 2014 Aug;42(4):1033-6. doi: 10.1042/BST20140096. Biochem Soc Trans. 2014. PMID: 25109998 Free PMC article. Review.
-
The coenzyme A biosynthetic pathway: A new tool for prodrug bioactivation.Arch Biochem Biophys. 2019 Sep 15;672:108069. doi: 10.1016/j.abb.2019.108069. Epub 2019 Aug 9. Arch Biochem Biophys. 2019. PMID: 31404525 Review.
Cited by
-
In Silico Approach: Anti-Tuberculosis Activity of Caespitate in the H37Rv Strain.Curr Issues Mol Biol. 2024 Jun 27;46(7):6489-6507. doi: 10.3390/cimb46070387. Curr Issues Mol Biol. 2024. PMID: 39057029 Free PMC article.
-
Structural and biochemical characterization of compounds inhibiting Mycobacterium tuberculosis pantothenate kinase.J Biol Chem. 2013 Jun 21;288(25):18260-70. doi: 10.1074/jbc.M113.476473. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661699 Free PMC article.
-
PanG, a new ketopantoate reductase involved in pantothenate synthesis.J Bacteriol. 2013 Mar;195(5):965-76. doi: 10.1128/JB.01740-12. Epub 2012 Dec 14. J Bacteriol. 2013. PMID: 23243306 Free PMC article.
-
Characterization and validation of Entamoeba histolytica pantothenate kinase as a novel anti-amebic drug target.Int J Parasitol Drugs Drug Resist. 2018 Apr;8(1):125-136. doi: 10.1016/j.ijpddr.2018.02.004. Epub 2018 Mar 1. Int J Parasitol Drugs Drug Resist. 2018. PMID: 29518650 Free PMC article.
-
Regulation of Coenzyme A Biosynthesis in the Hyperthermophilic Bacterium Thermotoga maritima.J Bacteriol. 2016 Jun 27;198(14):1993-2000. doi: 10.1128/JB.00077-16. Print 2016 Jul 15. J Bacteriol. 2016. PMID: 27161115 Free PMC article.
References
-
- Aceti, D. J., and J. G. Ferry. 1988. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. Evidence for regulation of synthesis. J. Biol. Chem. 263:15444-15448. - PubMed
-
- Aleshin, A. E., C. Kirby, X. Liu, G. P. Bourenkov, H. D. Bartunik, H. J. Fromm, and R. B. Honzatko. 2000. Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation. J. Mol. Biol. 296:1001-1015. - PubMed
-
- Arora, K. K., C. R. Filburn, and P. L. Pedersen. 1991. Glucose phosphorylation. Site-directed mutations which impair the catalytic function of hexokinase. J. Biol. Chem. 266:5359-5362. - PubMed
-
- Becker, D., M. Selbach, C. Rollenhagen, M. Ballmaier, T. F. Meyer, M. Mann, and D. Bumann. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303-307. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

