Structural parameters affecting the kinetics of RNA hairpin formation
- PMID: 16855293
- PMCID: PMC1524914
- DOI: 10.1093/nar/gkl445
Structural parameters affecting the kinetics of RNA hairpin formation
Abstract
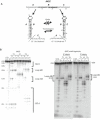
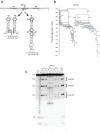
There is little experimental knowledge on the sequence dependent rate of hairpin formation in RNA. We have therefore designed RNA sequences that can fold into either of two mutually exclusive hairpins and have determined the ratio of folding of the two conformations, using structure probing. This folding ratio reflects their respective folding rates. Changing one of the two loop sequences from a purine- to a pyrimidine-rich loop did increase its folding rate, which corresponds well with similar observations in DNA hairpins. However, neither changing one of the loops from a regular non-GNRA tetra-loop into a stable GNRA tetra-loop, nor increasing the loop size from 4 to 6 nt did affect the folding rate. The folding kinetics of these RNAs have also been simulated with the program 'Kinfold'. These simulations were in agreement with the experimental results if the additional stabilization energies for stable tetra-loops were not taken into account. Despite the high stability of the stable tetra-loops, they apparently do not affect folding kinetics of these RNA hairpins. These results show that it is possible to experimentally determine relative folding rates of hairpins and to use these data to improve the computer-assisted simulation of the folding kinetics of stem-loop structures.
Figures
Similar articles
-
A thermodynamic study of unusually stable RNA and DNA hairpins.Nucleic Acids Res. 1991 Nov 11;19(21):5901-5. doi: 10.1093/nar/19.21.5901. Nucleic Acids Res. 1991. PMID: 1719483 Free PMC article.
-
Effect of loop composition on the stability and folding kinetics of RNA hairpins with large loops.Biochemistry. 2015 Mar 17;54(10):1886-96. doi: 10.1021/bi5014276. Epub 2015 Mar 4. Biochemistry. 2015. PMID: 25697574
-
RNA simulations: probing hairpin unfolding and the dynamics of a GNRA tetraloop.J Mol Biol. 2002 Apr 5;317(4):493-506. doi: 10.1006/jmbi.2002.5447. J Mol Biol. 2002. PMID: 11955005
-
Structures, kinetics, thermodynamics, and biological functions of RNA hairpins.Annu Rev Phys Chem. 2008;59:79-103. doi: 10.1146/annurev.physchem.59.032607.093743. Annu Rev Phys Chem. 2008. PMID: 17937599 Review.
-
Exceptionally stable nucleic acid hairpins.Annu Rev Biophys Biomol Struct. 1995;24:379-404. doi: 10.1146/annurev.bb.24.060195.002115. Annu Rev Biophys Biomol Struct. 1995. PMID: 7545040 Review.
Cited by
-
Predicting secondary structural folding kinetics for nucleic acids.Biophys J. 2010 Apr 21;98(8):1617-25. doi: 10.1016/j.bpj.2009.12.4319. Biophys J. 2010. PMID: 20409482 Free PMC article.
-
Probing the structural hierarchy and energy landscape of an RNA T-loop hairpin.Nucleic Acids Res. 2007;35(20):6995-7002. doi: 10.1093/nar/gkm719. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940098 Free PMC article.
-
RNA folding: conformational statistics, folding kinetics, and ion electrostatics.Annu Rev Biophys. 2008;37:197-214. doi: 10.1146/annurev.biophys.37.032807.125957. Annu Rev Biophys. 2008. PMID: 18573079 Free PMC article. Review.
-
Stem-loop structures can effectively substitute for an RNA pseudoknot in -1 ribosomal frameshifting.Nucleic Acids Res. 2011 Nov 1;39(20):8952-9. doi: 10.1093/nar/gkr579. Epub 2011 Jul 29. Nucleic Acids Res. 2011. PMID: 21803791 Free PMC article.
-
Utilizing RNA-seq Data to Infer Bacterial Transcription Termination Sites and Validate Predictions.Methods Mol Biol. 2024;2812:345-365. doi: 10.1007/978-1-0716-3886-6_19. Methods Mol Biol. 2024. PMID: 39068372
References
-
- Brion P., Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. - PubMed
-
- Gultyaev A.P., van Batenburg F.H., Pleij C.W.A. The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol. 1995;250:37–51. - PubMed
-
- Higgs P.G. Overlaps between RNA secondary structures. Phys. Rev. Lett. 1996;76:704–707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

