Binding of the COOH-terminal lysine residue of streptokinase to plasmin(ogen) kringles enhances formation of the streptokinase.plasmin(ogen) catalytic complexes
- PMID: 16857686
- PMCID: PMC2291350
- DOI: 10.1074/jbc.C600171200
Binding of the COOH-terminal lysine residue of streptokinase to plasmin(ogen) kringles enhances formation of the streptokinase.plasmin(ogen) catalytic complexes
Abstract
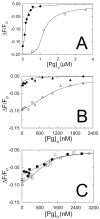
Streptokinase (SK) activates human fibrinolysis by inducing non-proteolytic activation of the serine proteinase zymogen, plasminogen (Pg), in the SK.Pg* catalytic complex. SK.Pg* proteolytically activates Pg to plasmin (Pm). SK-induced Pg activation is enhanced by lysine-binding site (LBS) interactions with kringles on Pg and Pm, as evidenced by inhibition of the reactions by the lysine analogue, 6-aminohexanoic acid. Equilibrium binding analysis and [Lys]Pg activation kinetics with wild-type SK, carboxypeptidase B-treated SK, and a COOH-terminal Lys414 deletion mutant (SKDeltaK414) demonstrated a critical role for Lys414 in the enhancement of [Lys]Pg and [Lys]Pm binding and conformational [Lys]Pg activation. The LBS-independent affinity of SK for [Glu]Pg was unaffected by deletion of Lys414. By contrast, removal of SK Lys414 caused 19- and 14-fold decreases in SK affinity for [Lys]Pg and [Lys]Pm binding in the catalytic mode, respectively. In kinetic studies of the coupled conformational and proteolytic activation of [Lys]Pg, SKDeltaK414 exhibited a corresponding 17-fold affinity decrease for formation of the SKDeltaK414.[Lys]Pg* complex. SKDeltaK414 binding to [Lys]Pg and [Lys]Pm and conformational [Lys]Pg activation were LBS-independent, whereas [Lys]Pg substrate binding and proteolytic [Lys]Pm generation remained LBS-dependent. We conclude that binding of SK Lys414 to [Lys]Pg and [Lys]Pm kringles enhances SK.[Lys]Pg* and SK.[Lys]Pm catalytic complex formation. This interaction is distinct structurally and functionally from LBS-dependent Pg substrate recognition by these complexes.
Figures



Similar articles
-
Plasminogen substrate recognition by the streptokinase-plasminogen catalytic complex is facilitated by Arg253, Lys256, and Lys257 in the streptokinase beta-domain and kringle 5 of the substrate.J Biol Chem. 2009 Jul 17;284(29):19511-21. doi: 10.1074/jbc.M109.005512. Epub 2009 May 27. J Biol Chem. 2009. PMID: 19473980 Free PMC article.
-
Rapid binding of plasminogen to streptokinase in a catalytic complex reveals a three-step mechanism.J Biol Chem. 2014 Oct 3;289(40):28006-18. doi: 10.1074/jbc.M114.589077. Epub 2014 Aug 19. J Biol Chem. 2014. PMID: 25138220 Free PMC article.
-
Role of the streptokinase alpha-domain in the interactions of streptokinase with plasminogen and plasmin.J Biol Chem. 2005 Mar 4;280(9):7504-10. doi: 10.1074/jbc.M411637200. Epub 2004 Dec 28. J Biol Chem. 2005. PMID: 15623524 Free PMC article.
-
Streptokinase binds preferentially to the extended conformation of plasminogen through lysine binding site and catalytic domain interactions.Biochemistry. 2000 Nov 14;39(45):13974-81. doi: 10.1021/bi000594i. Biochemistry. 2000. PMID: 11076540
-
Pathogen activators of plasminogen.J Thromb Haemost. 2015 Jun;13 Suppl 1(0 1):S106-14. doi: 10.1111/jth.12939. J Thromb Haemost. 2015. PMID: 26149011 Free PMC article. Review.
Cited by
-
Specificity of binding of the low density lipoprotein receptor-related protein to different conformational states of the clade E serpins plasminogen activator inhibitor-1 and proteinase nexin-1.J Biol Chem. 2009 Jul 3;284(27):17989-97. doi: 10.1074/jbc.M109.009530. Epub 2009 May 13. J Biol Chem. 2009. PMID: 19439404 Free PMC article.
-
Skizzle is a novel plasminogen- and plasmin-binding protein from Streptococcus agalactiae that targets proteins of human fibrinolysis to promote plasmin generation.J Biol Chem. 2010 Jul 2;285(27):21153-64. doi: 10.1074/jbc.M110.107730. Epub 2010 Apr 30. J Biol Chem. 2010. PMID: 20435890 Free PMC article.
-
In Vivo Tracking of Streptococcal Infections of Subcutaneous Origin in a Murine Model.Mol Imaging Biol. 2015 Dec;17(6):793-801. doi: 10.1007/s11307-015-0856-2. Mol Imaging Biol. 2015. PMID: 25921659 Free PMC article.
-
Proteases: Pivot Points in Functional Proteomics.Methods Mol Biol. 2019;1871:313-392. doi: 10.1007/978-1-4939-8814-3_20. Methods Mol Biol. 2019. PMID: 30276748 Free PMC article. Review.
-
Carbon and amide detect backbone assignment methods of a novel repeat protein from the staphylocoagulase in S. aureus.Biomol NMR Assign. 2017 Oct;11(2):243-249. doi: 10.1007/s12104-017-9757-4. Epub 2017 Aug 17. Biomol NMR Assign. 2017. PMID: 28819722 Free PMC article.
References
-
- Collen D, Lijnen HR. Blood. 1991;78:3114–3124. - PubMed
-
- McClintock DK, Bell PH. Biochem Biophys Res Commun. 1971;43:694–702. - PubMed
-
- Reddy KN, Markus G. J Biol Chem. 1972;247:1683–1691. - PubMed
-
- Schick LA, Castellino FJ. Biochem Biophys Res Commun. 1974;57:47–54. - PubMed
-
- Boxrud PD, Verhamme IM, Bock PE. J Biol Chem. 2004;279:36633–36641. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

