Aneuploidy and isochromosome formation in drug-resistant Candida albicans
- PMID: 16857942
- PMCID: PMC1717021
- DOI: 10.1126/science.1128242
Aneuploidy and isochromosome formation in drug-resistant Candida albicans
Abstract
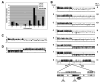
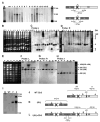
Resistance to the limited number of available antifungal drugs is a serious problem in the treatment of Candida albicans. We found that aneuploidy in general and a specific segmental aneuploidy, consisting of an isochromosome composed of the two left arms of chromosome 5, were associated with azole resistance. The isochromosome forms around a single centromere flanked by an inverted repeat and was found as an independent chromosome or fused at the telomere to a full-length homolog of chromosome 5. Increases and decreases in drug resistance were strongly associated with gain and loss of this isochromosome, which bears genes expressing the enzyme in the ergosterol pathway targeted by azole drugs, efflux pumps, and a transcription factor that positively regulates a subset of efflux pump genes.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

