Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage
- PMID: 16858410
- PMCID: PMC1523172
- DOI: 10.1038/sj.emboj.7601219
Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage
Abstract
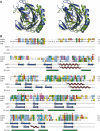
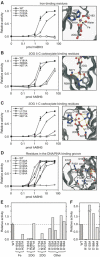
Methylating agents are ubiquitous in the environment, and central in cancer therapy. The 1-methyladenine and 3-methylcytosine lesions in DNA/RNA contribute to the cytotoxicity of such agents. These lesions are directly reversed by ABH3 (hABH3) in humans and AlkB in Escherichia coli. Here, we report the structure of the hABH3 catalytic core in complex with iron and 2-oxoglutarate (2OG) at 1.5 A resolution and analyse key site-directed mutants. The hABH3 structure reveals the beta-strand jelly-roll fold that coordinates a catalytically active iron centre by a conserved His1-X-Asp/Glu-X(n)-His2 motif. This experimentally establishes hABH3 as a structural member of the Fe(II)/2OG-dependent dioxygenase superfamily, which couples substrate oxidation to conversion of 2OG into succinate and CO2. A positively charged DNA/RNA binding groove indicates a distinct nucleic acid binding conformation different from that predicted in the AlkB structure with three nucleotides. These results uncover previously unassigned key catalytic residues, identify a flexible hairpin involved in nucleotide flipping and ss/ds-DNA discrimination, and reveal self-hydroxylation of an active site leucine that may protect against uncoupled generation of dangerous oxygen radicals.
Figures
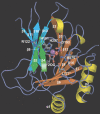


References
-
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 - PubMed
-
- Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell 92: 117–129 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

