Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging
- PMID: 16859491
- PMCID: PMC1698604
- DOI: 10.1042/BJ20060874
Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging
Abstract
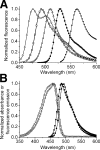
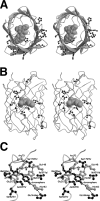
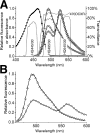
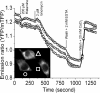
The arsenal of engineered variants of the GFP [green FP (fluorescent protein)] from Aequorea jellyfish provides researchers with a powerful set of tools for use in biochemical and cell biology research. The recent discovery of diverse FPs in Anthozoa coral species has provided protein engineers with an abundance of alternative progenitor FPs from which improved variants that complement or supersede existing Aequorea GFP variants could be derived. Here, we report the engineering of the first monomeric version of the tetrameric CFP (cyan FP) cFP484 from Clavularia coral. Starting from a designed synthetic gene library with mammalian codon preferences, we identified dimeric cFP484 variants with fluorescent brightness significantly greater than the wild-type protein. Following incorporation of dimer-breaking mutations and extensive directed evolution with selection for blue-shifted emission, high fluorescent brightness and photostability, we arrived at an optimized variant that we have named mTFP1 [monomeric TFP1 (teal FP 1)]. The new mTFP1 is one of the brightest and most photostable FPs reported to date. In addition, the fluorescence is insensitive to physiologically relevant pH changes and the fluorescence lifetime decay is best fitted as a single exponential. The 1.19 A crystal structure (1 A=0.1 nm) of mTFP1 confirms the monomeric structure and reveals an unusually distorted chromophore conformation. As we experimentally demonstrate, the high quantum yield of mTFP1 (0.85) makes it particularly suitable as a replacement for ECFP (enhanced CFP) or Cerulean as a FRET (fluorescence resonance energy transfer) donor to either a yellow or orange FP acceptor.
Figures



References
-
- Tsien R. Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. - PubMed
-
- Matz M. V., Fradkov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., Markelov M. L., Lukyanov S. A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. - PubMed
-
- Verkhusha V. V., Lukyanov K. A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004;22:289–296. - PubMed
-
- Zhang J., Campbell R. E., Ting A. Y., Tsien R. Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. - PubMed
-
- Chudakov D. M., Lukyanov S., Lukyanov K. A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

