yrGATE: a web-based gene-structure annotation tool for the identification and dissemination of eukaryotic genes
- PMID: 16859520
- PMCID: PMC1779557
- DOI: 10.1186/gb-2006-7-7-r58
yrGATE: a web-based gene-structure annotation tool for the identification and dissemination of eukaryotic genes
Abstract
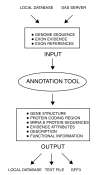
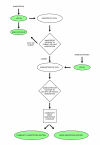
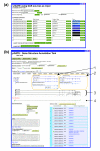
Your Gene structure Annotation Tool for Eukaryotes (yrGATE) provides an Annotation Tool and Community Utilities for worldwide web-based community genome and gene annotation. Annotators can evaluate gene structure evidence derived from multiple sources to create gene structure annotations. Administrators regulate the acceptance of annotations into published gene sets. yrGATE is designed to facilitate rapid and accurate annotation of emerging genomes as well as to confirm, refine, or correct currently published annotations. yrGATE is highly portable and supports different standard input and output formats. The yrGATE software and usage cases are available at http://www.plantgdb.org/prj/yrGATE.
Figures


Similar articles
-
GeneSeqer@PlantGDB: Gene structure prediction in plant genomes.Nucleic Acids Res. 2003 Jul 1;31(13):3597-600. doi: 10.1093/nar/gkg533. Nucleic Acids Res. 2003. PMID: 12824374 Free PMC article.
-
GASS: genome structural annotation for Eukaryotes based on species similarity.BMC Genomics. 2015 Mar 4;16(1):150. doi: 10.1186/s12864-015-1353-3. BMC Genomics. 2015. PMID: 25764973 Free PMC article.
-
PlantGDB: a resource for comparative plant genomics.Nucleic Acids Res. 2008 Jan;36(Database issue):D959-65. doi: 10.1093/nar/gkm1041. Epub 2007 Dec 6. Nucleic Acids Res. 2008. PMID: 18063570 Free PMC article.
-
Plant Gene and Alternatively Spliced Variant Annotator. A plant genome annotation pipeline for rice gene and alternatively spliced variant identification with cross-species expressed sequence tag conservation from seven plant species.Plant Physiol. 2007 Mar;143(3):1086-95. doi: 10.1104/pp.106.092460. Epub 2007 Jan 12. Plant Physiol. 2007. PMID: 17220363 Free PMC article.
-
Gene space completeness in complex plant genomes.Curr Opin Plant Biol. 2019 Apr;48:9-17. doi: 10.1016/j.pbi.2019.01.001. Epub 2019 Feb 21. Curr Opin Plant Biol. 2019. PMID: 30797187 Review.
Cited by
-
ORCAE: online resource for community annotation of eukaryotes.Nat Methods. 2012 Nov;9(11):1041. doi: 10.1038/nmeth.2242. Nat Methods. 2012. PMID: 23132114 No abstract available.
-
EuCAP, a Eukaryotic Community Annotation Package, and its application to the rice genome.BMC Genomics. 2007 Oct 25;8:388. doi: 10.1186/1471-2164-8-388. BMC Genomics. 2007. PMID: 17961238 Free PMC article.
-
MaizeGDB becomes 'sequence-centric'.Database (Oxford). 2009;2009:bap020. doi: 10.1093/database/bap020. Epub 2009 Dec 7. Database (Oxford). 2009. PMID: 21847242 Free PMC article.
-
Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing.BMC Genomics. 2007 Sep 19;8:330. doi: 10.1186/1471-2164-8-330. BMC Genomics. 2007. PMID: 17880721 Free PMC article.
-
Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize.PLoS One. 2011 Mar 10;6(3):e17855. doi: 10.1371/journal.pone.0017855. PLoS One. 2011. PMID: 21423772 Free PMC article.
References
-
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

