What makes species unique? The contribution of proteins with obscure features
- PMID: 16859532
- PMCID: PMC1779552
- DOI: 10.1186/gb-2006-7-7-r57
What makes species unique? The contribution of proteins with obscure features
Abstract
Background: Proteins with obscure features (POFs), which lack currently defined motifs or domains, represent between 18% and 38% of a typical eukaryotic proteome. To evaluate the contribution of this class of proteins to the diversity of eukaryotes, we performed a comparative analysis of the predicted proteomes derived from 10 different sequenced genomes, including budding and fission yeast, worm, fly, mosquito, Arabidopsis, rice, mouse, rat, and human.
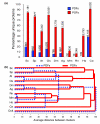
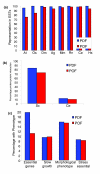
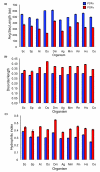
Results: Only 1,650 protein groups were found to be conserved among these proteomes (BLAST E-value threshold of 10(-6)). Of these, only three were designated as POFs. Surprisingly, we found that, on average, 60% of the POFs identified in these 10 proteomes (44,236 in total) were species specific. In contrast, only 7.5% of the proteins with defined features (PDFs) were species specific (17,554 in total). As a group, POFs appear similar to PDFs in their relative contribution to biological functions, as indicated by their expression, participation in protein-protein interactions and association with mutant phenotypes. However, POF have more predicted disordered structure than PDFs, implying that they may exhibit preferential involvement in species-specific regulatory and signaling networks.
Conclusion: Because the majority of eukaryotic POFs are not well conserved, and by definition do not have defined domains or motifs upon which to formulate a functional working hypothesis, understanding their biochemical and biological functions will require species-specific investigations.
Figures


Similar articles
-
POFs: what we don't know can hurt us.Trends Plant Sci. 2007 Nov;12(11):492-496. doi: 10.1016/j.tplants.2007.08.018. Epub 2007 Oct 24. Trends Plant Sci. 2007. PMID: 17928258
-
C-terminal motif prediction in eukaryotic proteomes using comparative genomics and statistical over-representation across protein families.BMC Genomics. 2007 Jun 26;8:191. doi: 10.1186/1471-2164-8-191. BMC Genomics. 2007. PMID: 17594486 Free PMC article.
-
Comparative proteomics uncovers the signature of natural selection acting on the ejaculate proteomes of two cricket species isolated by postmating, prezygotic phenotypes.Mol Biol Evol. 2011 Jan;28(1):423-35. doi: 10.1093/molbev/msq230. Epub 2010 Aug 30. Mol Biol Evol. 2011. PMID: 20805188
-
Genome-scale prediction of proteins with long intrinsically disordered regions.Proteins. 2014 Jan;82(1):145-58. doi: 10.1002/prot.24348. Epub 2013 Sep 17. Proteins. 2014. PMID: 23798504
-
Trends in genome dynamics among major orders of insects revealed through variations in protein families.BMC Genomics. 2015 Aug 7;16(1):583. doi: 10.1186/s12864-015-1771-2. BMC Genomics. 2015. PMID: 26251035 Free PMC article.
Cited by
-
JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom.Nucleic Acids Res. 2023 Sep 8;51(16):8383-8401. doi: 10.1093/nar/gkad616. Nucleic Acids Res. 2023. PMID: 37526283 Free PMC article.
-
Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function.Plant Physiol. 2008 Sep;148(1):280-92. doi: 10.1104/pp.108.124875. Epub 2008 Jul 9. Plant Physiol. 2008. PMID: 18614705 Free PMC article.
-
Characterization of the ERP gene family in Arabidopsis thaliana.Plant Signal Behav. 2021 Jun 3;16(6):1913301. doi: 10.1080/15592324.2021.1913301. Epub 2021 Apr 27. Plant Signal Behav. 2021. PMID: 33906568 Free PMC article.
-
Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.).J Exp Bot. 2010;61(1):143-56. doi: 10.1093/jxb/erp289. J Exp Bot. 2010. PMID: 19858118 Free PMC article.
-
Unique genes in plants: specificities and conserved features throughout evolution.BMC Evol Biol. 2008 Oct 10;8:280. doi: 10.1186/1471-2148-8-280. BMC Evol Biol. 2008. PMID: 18847470 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

