Tertiary contacts distant from the active site prime a ribozyme for catalysis
- PMID: 16859740
- PMCID: PMC4447102
- DOI: 10.1016/j.cell.2006.06.036
Tertiary contacts distant from the active site prime a ribozyme for catalysis
Abstract
Minimal hammerhead ribozymes have been characterized extensively by static and time-resolved crystallography as well as numerous biochemical analyses, leading to mutually contradictory mechanistic explanations for catalysis. We present the 2.2 A resolution crystal structure of a full-length Schistosoma mansoni hammerhead ribozyme that permits us to explain the structural basis for its 1000-fold catalytic enhancement. The full-length hammerhead structure reveals how tertiary interactions occurring remotely from the active site prime this ribozyme for catalysis. G-12 and G-8 are positioned consistent with their previously suggested roles in acid-base catalysis, the nucleophile is aligned with a scissile phosphate positioned proximal to the A-9 phosphate, and previously unexplained roles of other conserved nucleotides become apparent within the context of a distinctly new fold that nonetheless accommodates the previous structural studies. These interactions permit us to explain the previously irreconcilable sets of experimental results in a unified, consistent, and unambiguous manner.
Figures
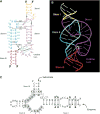




References
-
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. - PubMed
-
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. - PubMed
-
- Clouet-d’Orval B, Uhlenbeck OC. Hammerhead ribozymes with a faster cleavage rate. Biochemistry. 1997;36:9087–9092. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

