Interaction of protegrin-1 with lipid bilayers: membrane thinning effect
- PMID: 16861271
- PMCID: PMC1578484
- DOI: 10.1529/biophysj.106.084046
Interaction of protegrin-1 with lipid bilayers: membrane thinning effect
Abstract
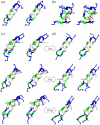
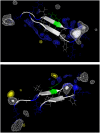
Protegrins (PG) are important in defending host tissues, preventing infection via an attack on the membrane surface of invading microorganisms. Protegrins have powerful antibiotic abilities, but the molecular-level mechanisms underlying the interactions of their beta-sheet motifs with the membrane are not known. Protegrin-1 (PG-1) is composed of 18 amino acids with a high content of basic residues and two disulfide bonds. Here we focused on the stability of PG-1 at the amphipathic interface in lipid bilayers and on the details of the peptide-membrane interactions. We simulated all-atom models of the PG-1 monomer with explicit water and lipid bilayers composed of both homogeneous POPC (palmitoyl-oleyl-phosphatidylcholine) lipids and a mixture of POPC/POPG (palmitoyl-oleyl-phosphatidylglycerol) (4:1) lipids. We observed that local thinning of the lipid bilayers mediated by the peptide is enhanced in the lipid bilayer containing POPG, consistent with experimental results of selective membrane targeting. The beta-hairpin motif of PG-1 is conserved in both lipid settings, whereas it is highly bent in aqueous solution. The conformational dynamics of PG-1, especially the highly charged beta-hairpin turn region, are found to be mostly responsible for disturbing the membrane. Even though the eventual membrane disruption requires PG-1 oligomers, our simulations clearly show the first step of the monomeric effects. The thinning effects in the bilayer should relate to pore/channel formation in the lipid bilayer and thus be responsible for further defects in the membrane caused by oligomer.
Figures







References
-
- Kourie, J. I., and A. A. Shorthouse. 2000. Properties of cytotoxic peptide-formed ion channels. Am. J. Physiol. Cell Physiol. 278:C1063–C1087. - PubMed
-
- Lai, J. R., B. R. Huck, B. Weisblum, and S. H. Gellman. 2002. Design of non-cysteine-containing antimicrobial β-hairpins: structure-activity relationship studies with linear protegrin-1 analogues. Biochemistry. 41:12835–12842. - PubMed
-
- Gobbo, M., L. Biondi, F. Filira, R. Gennaro, M. Benincasa, B. Scolaro, and R. Rocchi. 2002. Antimicrobial peptides: synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1b analogues. J. Med. Chem. 45:4494–4504. - PubMed
-
- Panchal, R. G., M. L. Smart, D. N. Bowser, D. A. Williams, and S. Petrou. 2002. Pore-forming proteins and their application in biotechnology. Curr. Pharm. Biotechnol. 3:99–115. - PubMed
-
- Sitaram, N., and R. Nagaraj. 1999. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochim. Biophys. Acta. 1462:29–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

