Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli
- PMID: 16861298
- PMCID: PMC1544242
- DOI: 10.1073/pnas.0604744103
Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli
Abstract
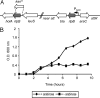
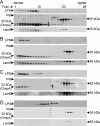
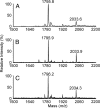
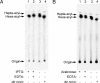
The outer membrane of most Gram-negative bacteria is made up of LPS, and in nearly all bacteria that contain LPS it is essential for the life of the organism. The lipid portion of this molecule, lipid A, also known as endotoxin, is a potent activator of the innate immune response. More than 50 genes are required to synthesize LPS and assemble it at the cell surface. Enormous progress has been made in elucidating the structure and biosynthesis of LPS, but until recently the cellular components required for its transport from its site of synthesis in the inner membrane to its final cellular location at the cell surface remained elusive. Here we describe the identification of a protein complex that functions to assemble LPS at the surface of the cell. This complex contains two proteins: Imp, already identified as an essential outer-membrane protein implicated in LPS assembly; and another protein, RlpB, heretofore identified only as a rare lipoprotein. We show that RlpB is also essential for cell viability and that the Imp/RlpB complex is responsible for LPS reaching the outer surface of the outer membrane.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

