STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes
- PMID: 16861352
- PMCID: PMC1895864
- DOI: 10.1182/blood-2005-08-007377
STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes
Abstract
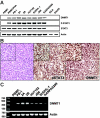
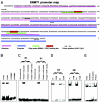
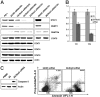
In this study, we demonstrated that STAT3, a well-characterized transcription factor expressed in continuously activated oncogenic form in the large spectrum of cancer types, induces in malignant T lymphocytes the expression of DNMT1, the key effector of epigenetic gene silencing. STAT3 binds in vitro to 2 STAT3 SIE/GAS-binding sites identified in promoter 1 and enhancer 1 of the DNMT1 gene. STAT3 also binds to the promoter 1 region and induces its activity in vivo. Treatment of the malignant T lymphocytes with STAT3 siRNA abrogates expression of DNMT1, inhibits cell growth, and induces programmed cell death. In turn, inhibition of DNMT1 by a small molecule inhibitor, 5-aza-2-deoxy-cytidine, and 2 DNMT1 antisense DNA oligonucleotides inhibits the phosphorylation of STAT3. These data indicate that STAT3 may in part transform cells by fostering epigenetic silencing of tumor-suppressor genes. They also indicate that by inducing DNMT1, STAT3 facilitates its own persistent activation in malignant T cells. Finally, these data provide further rationale for therapeutically targeting STAT3 in T-cell lymphomas and, possibly, other malignancies.
Figures


References
-
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349: 2042-2054. - PubMed
-
- Hodge DR, Peng B, Cherry JC, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promoter methylation. Cancer Res. 2005;65: 4673-4682. - PubMed
-
- Robertson KD. DNA methylation and chromatin—unraveling the tangled web. Oncogene. 2002;21: 5361-5379. - PubMed
-
- Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416: 552-556. - PubMed
-
- Szyf M. Utilization of antisense oligonucleotides to study the role of 5-cytosine DNA methyltransferase in cellular transformation and oncogenesis. Methods. 2002;27: 184-191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

