A genomic code for nucleosome positioning
- PMID: 16862119
- PMCID: PMC2623244
- DOI: 10.1038/nature04979
A genomic code for nucleosome positioning
Abstract
Eukaryotic genomes are packaged into nucleosome particles that occlude the DNA from interacting with most DNA binding proteins. Nucleosomes have higher affinity for particular DNA sequences, reflecting the ability of the sequence to bend sharply, as required by the nucleosome structure. However, it is not known whether these sequence preferences have a significant influence on nucleosome position in vivo, and thus regulate the access of other proteins to DNA. Here we isolated nucleosome-bound sequences at high resolution from yeast and used these sequences in a new computational approach to construct and validate experimentally a nucleosome-DNA interaction model, and to predict the genome-wide organization of nucleosomes. Our results demonstrate that genomes encode an intrinsic nucleosome organization and that this intrinsic organization can explain approximately 50% of the in vivo nucleosome positions. This nucleosome positioning code may facilitate specific chromosome functions including transcription factor binding, transcription initiation, and even remodelling of the nucleosomes themselves.
Figures
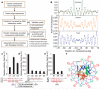


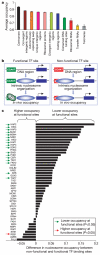
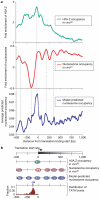
Comment in
-
Genomics: predictable packaging.Nature. 2006 Aug 17;442(7104):750-2. doi: 10.1038/442750a. Nature. 2006. PMID: 16915272 No abstract available.
References
-
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. - PubMed
-
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. - PubMed
-
- Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. - PubMed
-
- van Holde KE. Chromatin. Springer; New York: 1989.
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

