From functional genomics to functional immunomics: new challenges, old problems, big rewards
- PMID: 16863395
- PMCID: PMC1523295
- DOI: 10.1371/journal.pcbi.0020081
From functional genomics to functional immunomics: new challenges, old problems, big rewards
Abstract
The development of DNA microarray technology a decade ago led to the establishment of functional genomics as one of the most active and successful scientific disciplines today. With the ongoing development of immunomic microarray technology-a spatially addressable, large-scale technology for measurement of specific immunological response-the new challenge of functional immunomics is emerging, which bears similarities to but is also significantly different from functional genomics. Immunonic data has been successfully used to identify biological markers involved in autoimmune diseases, allergies, viral infections such as human immunodeficiency virus (HIV), influenza, diabetes, and responses to cancer vaccines. This review intends to provide a coherent vision of this nascent scientific field, and speculate on future research directions. We discuss at some length issues such as epitope prediction, immunomic microarray technology and its applications, and computation and statistical challenges related to functional immunomics. Based on the recent discovery of regulation mechanisms in T cell responses, we envision the use of immunomic microarrays as a tool for advances in systems biology of cellular immune responses, by means of immunomic regulatory network models.
Conflict of interest statement
Figures


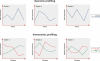
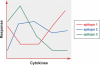

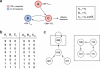
References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns via a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnol. 1996;14:1675–1680. - PubMed
-
- Chen Y, Dougherty E, Bittner M. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J Biomed Opt. 1997;2:364–374. - PubMed
-
- Kohane IS, Kho A, Butt AJ. Microarrays for an integrative genomics. Cambridge (Massachusetts): The MIT Press; 2002. 326. p.
-
- Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

