Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity
- PMID: 16864656
- PMCID: PMC2064238
- DOI: 10.1083/jcb.200604031
Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity
Abstract
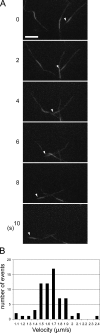
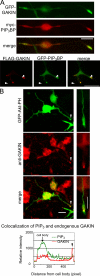
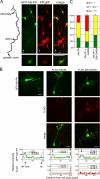
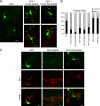
Phosphatidylinositol-(3,4,5)-trisphosphate (PIP3), a product of phosphatidylinositol 3-kinase, is an important second messenger implicated in signal transduction and membrane transport. In hippocampal neurons, the accumulation of PIP3 at the tip of neurite initiates the axon specification and neuronal polarity formation. We show that guanylate kinase-associated kinesin (GAKIN), a kinesin-like motor protein, directly interacts with a PIP3-interacting protein, PIP3BP, and mediates the transport of PIP3-containing vesicles. Recombinant GAKIN and PIP3BP form a complex on synthetic liposomes containing PIP3 and support the motility of the liposomes along microtubules in vitro. In PC12 cells and cultured hippocampal neurons, transport activity of GAKIN contributes to the accumulation of PIP3 at the tip of neurites. In hippocampal neurons, altered accumulation of PIP3 by overexpression of GAKIN constructs led to the loss of the axonally differentiated neurites. Together, these results suggest that, in neurons, the GAKIN-PIP3BP complex transports PIP3 to the neurite ends and regulates neuronal polarity formation.
Figures




References
-
- Asaba, N., T. Hanada, A. Takeuchi, and A.H. Chishti. 2003. Direct interaction with a kinesin-related motor mediates transport of mammalian discs large tumor suppressor homologue in epithelial cells. J. Biol. Chem. 278:8395–8400. - PubMed
-
- Banker, G., and K. Goslin. 1998. Culturing Nerve Cells. 2nd edition. The MIT Press, Cambridge, MA. 666 pp.
-
- Bourne, H.R., and O. Weiner. 2002. A chemical compass. Nature. 419:21. - PubMed
-
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

