Targeting lentiviral vectors to specific cell types in vivo
- PMID: 16864770
- PMCID: PMC1518805
- DOI: 10.1073/pnas.0604993103
Targeting lentiviral vectors to specific cell types in vivo
Abstract
We have developed an efficient method to target lentivirus-mediated gene transduction to a desired cell type. It involves incorporation of antibody and fusogenic protein as two distinct molecules into the lentiviral surface. The fusogen is constructed by modifying viral envelope proteins, so that they lack the ability to bind to their cognate receptor but still retain the ability to trigger pH-dependent membrane fusion. Thus, the specificity of such a lentiviral vector is solely determined by the antibody, which is chosen to recognize a specific surface antigen of the desired cell type. This specific binding then induces endocytosis of the surface antigen, bringing the lentivirus into an endosome. There, the fusogen responds to the low pH environment and mediates membrane fusion, allowing the virus core to enter the cytosol. Using CD20 as a target antigen for human B cells, we have demonstrated that this targeting strategy is effective both in vitro and in intact animals. This methodology is flexible and can be extended to other forms of cell type-specific recognition to mediate targeting. The only requirement is that the antibody (or other binding protein) must be endocytosed after interaction with its cell surface-binding determinant.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


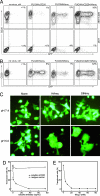

Similar articles
-
Visualization of targeted transduction by engineered lentiviral vectors.Gene Ther. 2008 Oct;15(20):1384-96. doi: 10.1038/gt.2008.87. Epub 2008 May 15. Gene Ther. 2008. PMID: 18480844 Free PMC article.
-
Gamma-retroviral vectors enveloped with an antibody and an engineered fusogenic protein achieved antigen-specific targeting.Biotechnol Bioeng. 2008 Oct 1;101(2):357-68. doi: 10.1002/bit.21903. Biotechnol Bioeng. 2008. PMID: 18435481 Free PMC article.
-
Redirecting lentiviral vectors by insertion of integrin-tageting peptides into envelope proteins.J Gene Med. 2009 Jul;11(7):549-58. doi: 10.1002/jgm.1339. J Gene Med. 2009. PMID: 19434609 Free PMC article.
-
Pseudotyped Lentiviral Vectors: One Vector, Many Guises.Hum Gene Ther Methods. 2017 Dec;28(6):291-301. doi: 10.1089/hgtb.2017.084. Epub 2017 Sep 4. Hum Gene Ther Methods. 2017. PMID: 28870117 Review.
-
Pseudotyping Lentiviral Vectors: When the Clothes Make the Virus.Viruses. 2020 Nov 16;12(11):1311. doi: 10.3390/v12111311. Viruses. 2020. PMID: 33207797 Free PMC article. Review.
Cited by
-
Targeting lentiviral vector to specific cell types through surface displayed single chain antibody and fusogenic molecule.Virol J. 2010 Feb 11;7:35. doi: 10.1186/1743-422X-7-35. Virol J. 2010. PMID: 20149250 Free PMC article.
-
miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer.Br J Cancer. 2010 Mar 2;102(5):883-91. doi: 10.1038/sj.bjc.6605570. Epub 2010 Feb 16. Br J Cancer. 2010. PMID: 20160723 Free PMC article.
-
Versatile targeting system for lentiviral vectors involving biotinylated targeting molecules.Virology. 2018 Dec;525:170-181. doi: 10.1016/j.virol.2018.09.017. Epub 2018 Oct 2. Virology. 2018. PMID: 30290312 Free PMC article.
-
Specific transduction of HIV-susceptible cells for CCR5 knockdown and resistance to HIV infection: a novel method for targeted gene therapy and intracellular immunization.J Acquir Immune Defic Syndr. 2009 Oct 1;52(2):152-61. doi: 10.1097/QAI.0b013e3181b010a0. J Acquir Immune Defic Syndr. 2009. PMID: 19593160 Free PMC article.
-
A versatile targeting system with lentiviral vectors bearing the biotin-adaptor peptide.J Gene Med. 2009 Aug;11(8):655-63. doi: 10.1002/jgm.1345. J Gene Med. 2009. PMID: 19455593 Free PMC article.
References
-
- Verma I. M., Somia N. Nature. 1997;389:239–242. - PubMed
-
- Somia N., Verma I. M. Nat. Rev. Genet. 2000;1:91–99. - PubMed
-
- Lavillette D., Russell S. J., Cosset F. L. Curr. Opin. Biotech. 2001;12:461–466. - PubMed
-
- Sandrin V., Russell S. J., Cosset F. L. Curr. Top. Microbiol. Immunol. 2003;281:137–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

