Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function
- PMID: 16864783
- PMCID: PMC1518800
- DOI: 10.1073/pnas.0603363103
Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function
Abstract
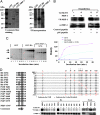
Cyclin D1 promotes nuclear DNA synthesis through phosphorylation and inactivation of the pRb tumor suppressor. Herein, cyclin D1 deficiency increased mitochondrial size and activity that was rescued by cyclin D1 in a Cdk-dependent manner. Nuclear respiratory factor 1 (NRF-1), which induces nuclear-encoded mitochondrial genes, was repressed in expression and activity by cyclin D1. Cyclin D1-dependent kinase phosphorylates NRF-1 at S47. Cyclin D1 abundance thus coordinates nuclear DNA synthesis and mitochondrial function.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Cyclin D1 determines mitochondrial function in vivo.Mol Cell Biol. 2006 Jul;26(14):5449-69. doi: 10.1128/MCB.02074-05. Mol Cell Biol. 2006. PMID: 16809779 Free PMC article.
-
Translokin (Cep57) interacts with cyclin D1 and prevents its nuclear accumulation in quiescent fibroblasts.Traffic. 2011 May;12(5):549-62. doi: 10.1111/j.1600-0854.2011.01176.x. Epub 2011 Mar 15. Traffic. 2011. PMID: 21306487
-
Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1.Oncogene. 2006 Oct 12;25(47):6291-303. doi: 10.1038/sj.onc.1209644. Epub 2006 May 29. Oncogene. 2006. PMID: 16732330
-
Mitochondrial reactive oxygen species-mediated genomic instability in low-dose irradiated human cells through nuclear retention of cyclin D1.Cell Cycle. 2016 Jun 2;15(11):1410-4. doi: 10.1080/15384101.2016.1170271. Epub 2016 Apr 14. Cell Cycle. 2016. PMID: 27078622 Free PMC article. Review.
-
New roles of cyclin D1.Am J Pathol. 2013 Jul;183(1):3-9. doi: 10.1016/j.ajpath.2013.03.001. Am J Pathol. 2013. PMID: 23790801 Free PMC article. Review.
Cited by
-
An analysis of cyclin D1, cytokeratin 5/6 and cytokeratin 8/18 expression in breast papillomas and papillary carcinomas.Diagn Pathol. 2013 Jan 18;8:8. doi: 10.1186/1746-1596-8-8. Diagn Pathol. 2013. PMID: 23327593 Free PMC article.
-
Cyclin D1 targets hexokinase 2 to control aerobic glycolysis in myeloma cells.Oncogenesis. 2020 Jul 24;9(7):68. doi: 10.1038/s41389-020-00253-3. Oncogenesis. 2020. PMID: 32709889 Free PMC article.
-
Two-way communication between the metabolic and cell cycle machineries: the molecular basis.Cell Cycle. 2015;14(13):2022-32. doi: 10.1080/15384101.2015.1044172. Cell Cycle. 2015. PMID: 26038996 Free PMC article. Review.
-
Small non-coding RNAs govern mammary gland tumorigenesis.J Mammary Gland Biol Neoplasia. 2012 Mar;17(1):59-64. doi: 10.1007/s10911-012-9246-4. Epub 2012 Mar 1. J Mammary Gland Biol Neoplasia. 2012. PMID: 22382486 Free PMC article. Review.
-
CDK2 regulates the NRF1/Ehmt1 axis during meiotic prophase I.J Cell Biol. 2019 Sep 2;218(9):2896-2918. doi: 10.1083/jcb.201903125. Epub 2019 Jul 26. J Cell Biol. 2019. PMID: 31350280 Free PMC article.
References
-
- Shadel G. S., Clayton D. A. Annu. Rev. Biochem. 1997;66:409–435. - PubMed
-
- Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., Clayton D. A. Nat. Genet. 1998;18:231–236. - PubMed
-
- Evans M. J., Scarpulla R. C. Genes Dev. 1990;4:1023–1034. - PubMed
-
- Scarpulla R. C. Biochim. Biophys. Acta. 2002;1576:1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

